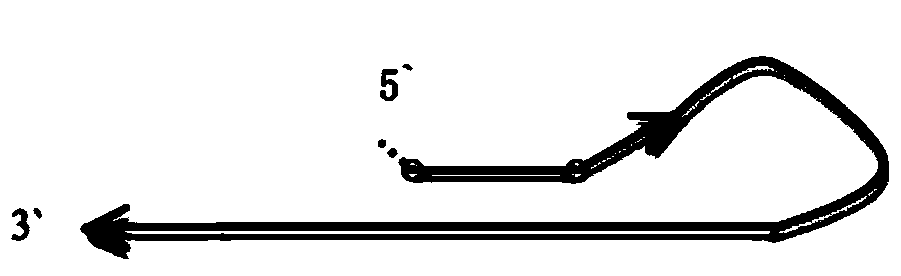
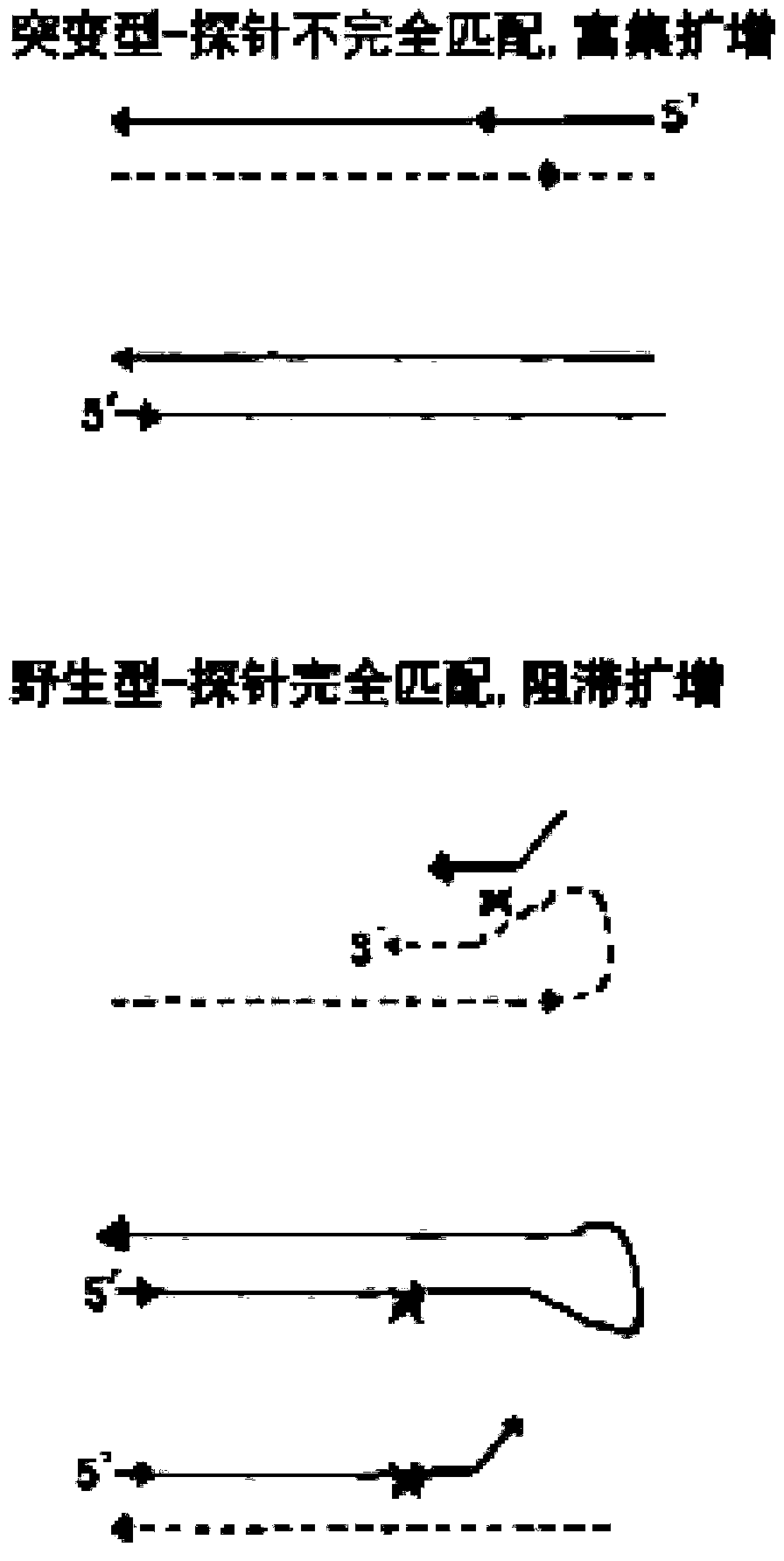
Low-frequency mutation detection method based on snapback primer technology
A technology of snapback probe primers and detection methods, which is applied in the field of gene detection, can solve problems such as obstacles, and achieve the effects of convenient operation, less pollution, and good enrichment and amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
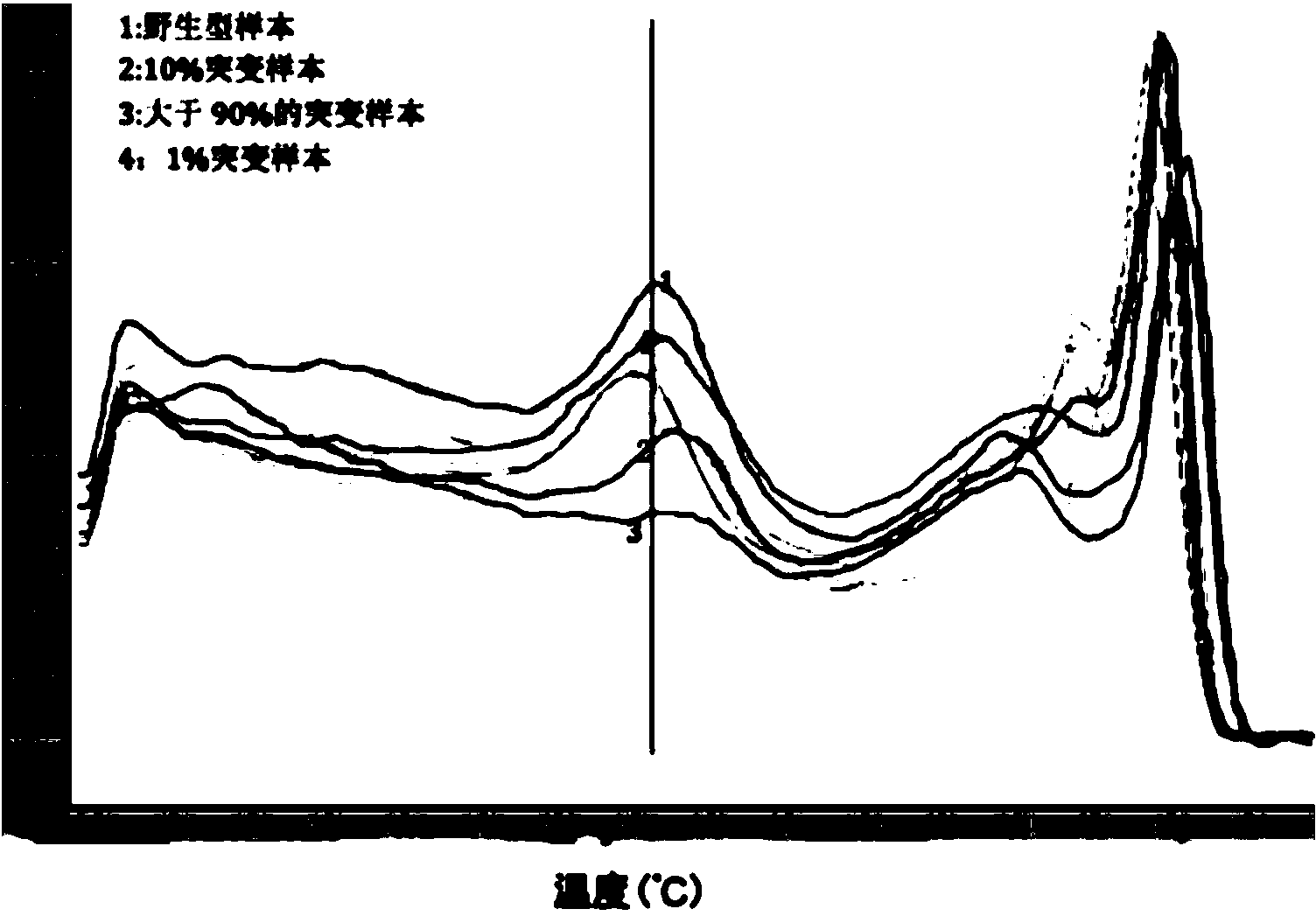
[0060] Use the EGFR gene exon 19 deletion mutation sample, mix it with normal human sample DNA according to different ratios, and make the mutation accounted for 1 / 106, 1 / 105, 1 / 104, 1 / 1000, 1 / 100, 1 / 10 The mixed mutant samples, pure, mutant, and wild-type samples are amplified using the rebound probe common amplification method, and the enrichment amplification method is used for amplification, respectively, HRM detection is performed, and the enriched amplified PCR products are sent to sequencing. Detection sensitivity, enrichment efficiency.
[0061] 1.1. Preparation of DNA template:
[0062] 1.1.1 Use QIAamp DNA Mini Kit (Qiagen, Germany) to extract DNA from adjacent tissues and tumor tissues of lung cancer patients, use Nano Drop 2000 spectrophotometer to detect the concentration, and dilute to 20ng / μl with sterile double-distilled water, and obtain by sanger sequencing Wild-type sample, mutant sample (15bp deletion).
[0063] 1.1.2 Mix and match the same concentration (20ng / μ...
Embodiment 2
[0081] Use the EGFR gene L858R site mutation sample, mix it with normal human sample DNA according to different proportions, and make the mutation account for 1 / 10 6 , 1 / 10 5 , 1 / 10 4 , 1 / 1000, 1 / 100, 1 / 10 ratios of mutant mixed samples, pure, mutant, and wild-type samples are amplified using the rebound probe common amplification method, and the enrichment amplification method is used for HRM detection. , And send the enriched and amplified PCR products for sequencing. Detection sensitivity, enrichment efficiency.
[0082] 2.1. Preparation of DNA template:
[0083] 2.1.1 Use QIAamp DNA Mini Kit (Qiagen, Germany) to extract DNA from adjacent tissues and tumor tissues of lung cancer patients, use Nano Drop 2000 spectrophotometer to detect the concentration, and dilute to 20ng / μl with sterile double-distilled water, and sequence by sanger Obtain wild-type samples, mutant samples (L858R).
[0084] 2.1.2 Mix equal concentrations (20ng / μl) of mutant and wild-type DNA according to the re...
Embodiment 3
[0104] Use KRAS gene locus mutation samples, mix with normal human sample DNA in different proportions, and make mutations account for 1 / 10 6 , 1 / 10 5 , 1 / 10 4 , 1 / 1000, 1 / 100, 1 / 10 ratios of mutant mixed samples, pure, mutant, and wild-type samples are amplified using the rebound probe common amplification method, and the enrichment amplification method is used for HRM detection. , And send the enriched and amplified PCR products for sequencing. Detection sensitivity, enrichment efficiency.
[0105] 3.1. Preparation of DNA template:
[0106] 3.1.1 Use QIAamp DNA Mini Kit (Qiagen, Germany) to extract DNA from adjacent tissues and tumor tissues of lung cancer patients, use Nano Drop 2000 spectrophotometer to detect the concentration, and dilute to 20ng / μl with sterile double-distilled water, and obtain by sanger sequencing Wild type sample, mutant sample (G12A).
[0107] 3.1.2 Mix the mutant and wild-type DNA with equal concentration (20ng / μl) according to the required ratio to obta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com