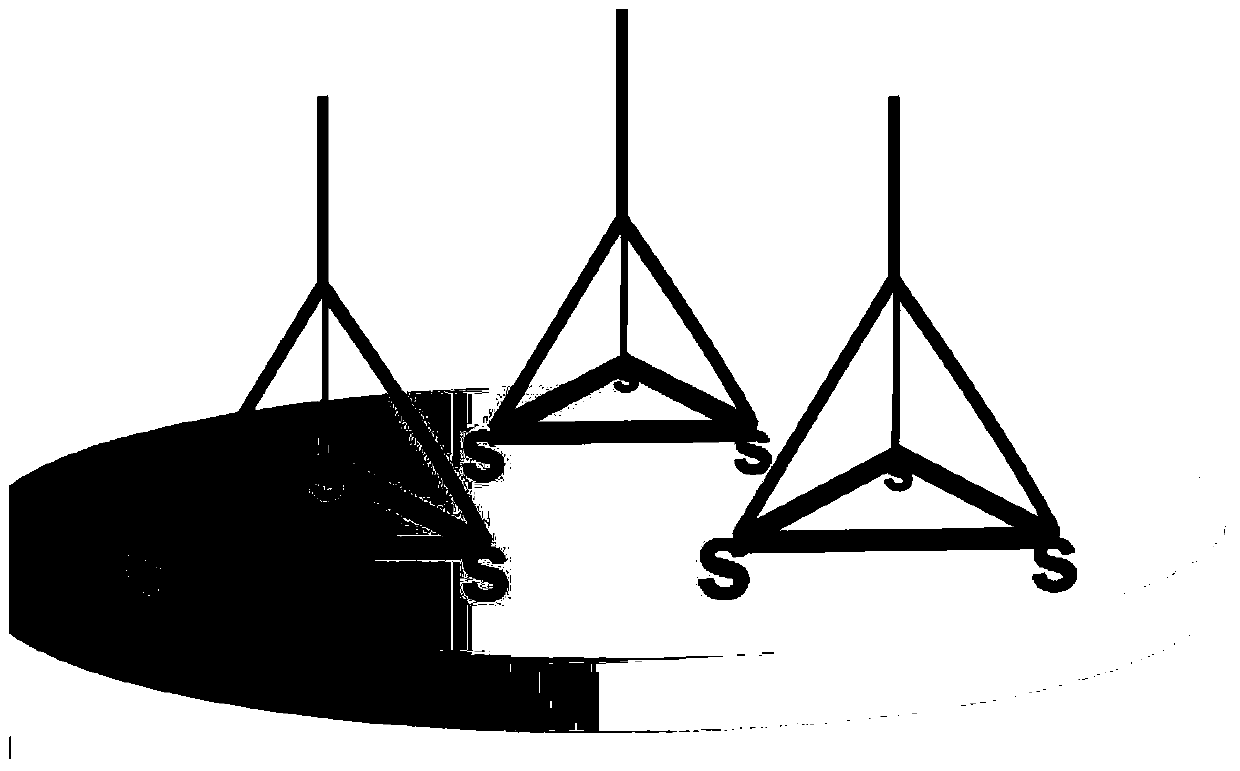
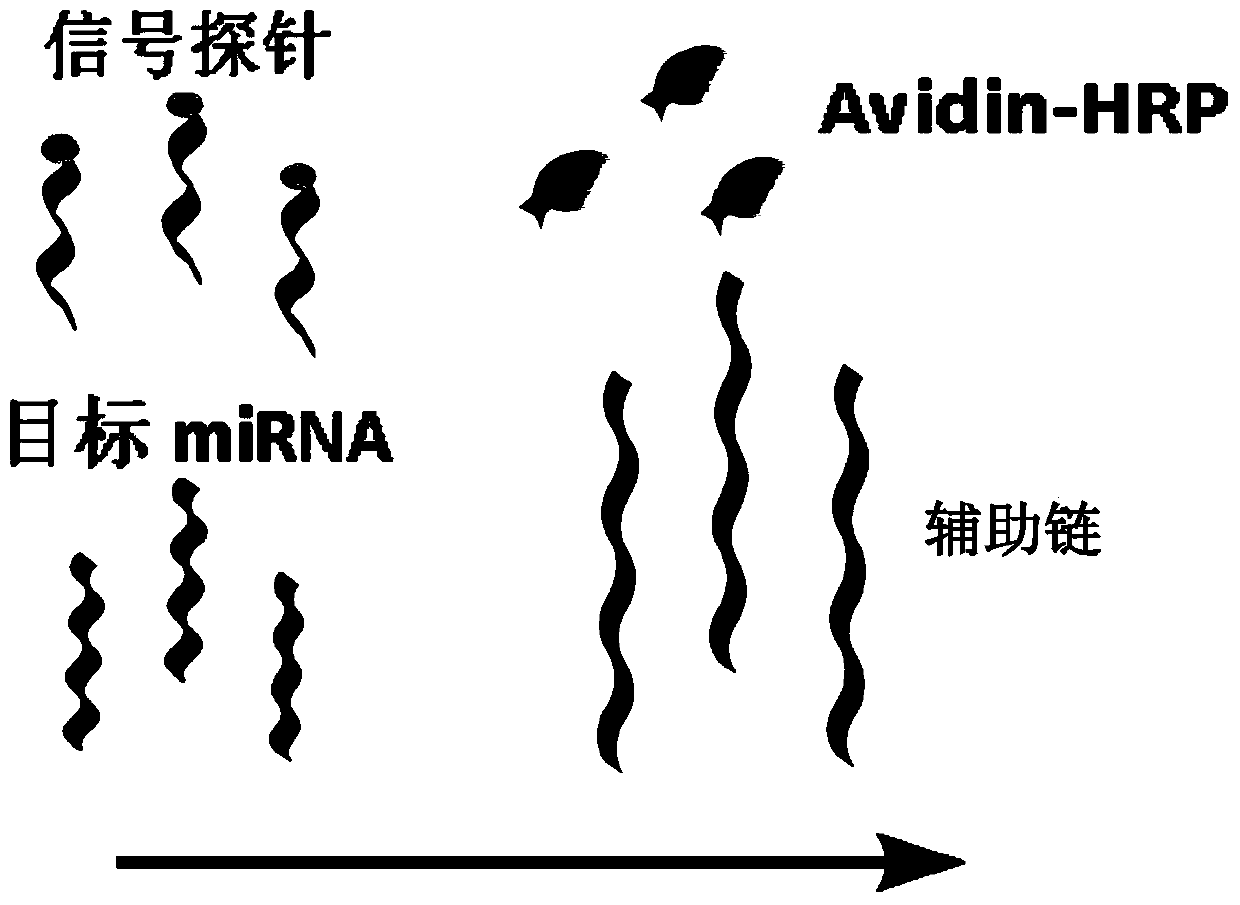
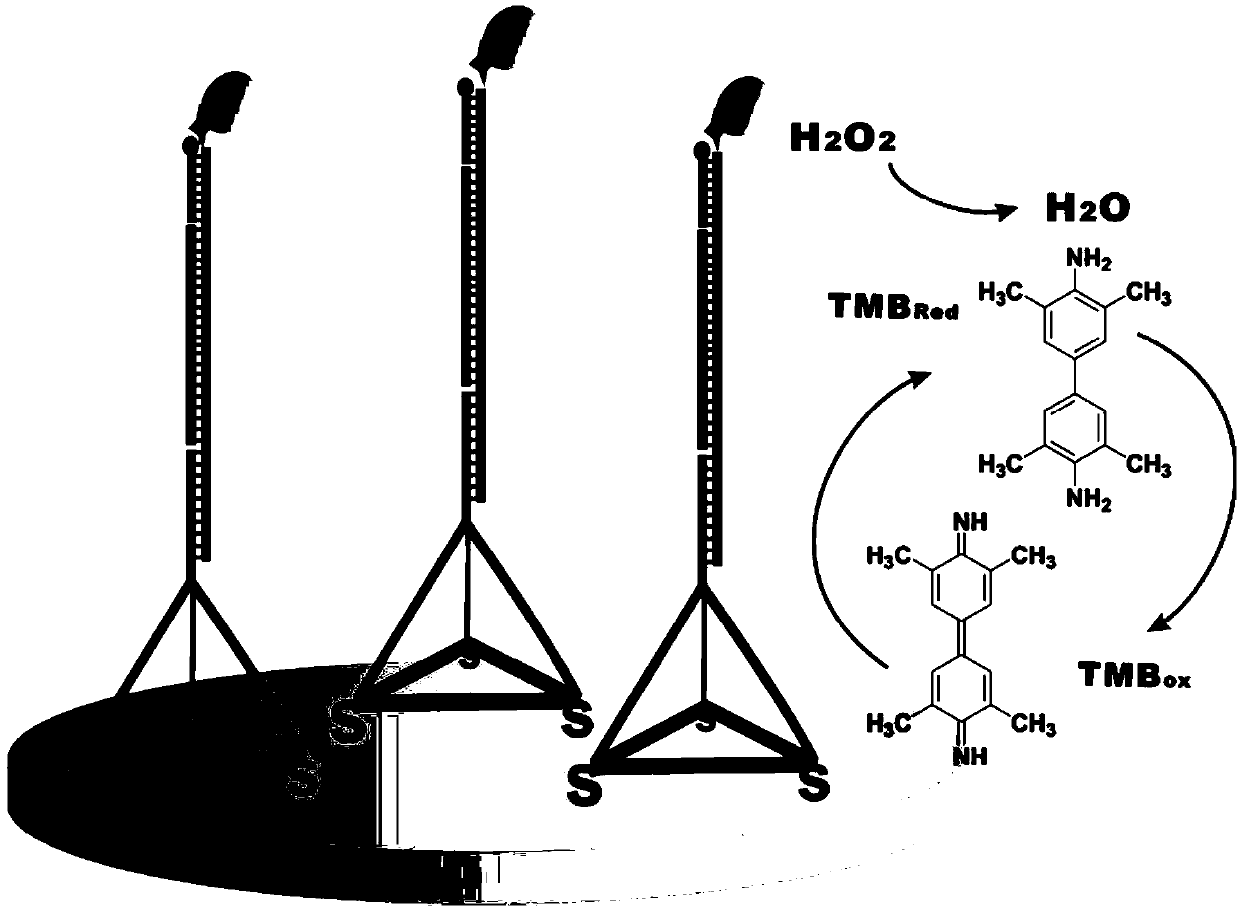
Method for detecting miRNA (ribonucleic acid)
A step-by-step, self-supporting technology that can be used in biochemical equipment and methods, measuring devices, and microbial determination/inspection, etc., and can solve the problems of long detection time, complicated operation and high cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] 1. Reagents and materials
[0042]For the convenience of description, the four ssDNA single strands whose sequences are shown in SEQ ID NO.1, SEQ ID NO.2, SEQ ID NO.3 and SEQ ID NO.4 are called Tetra-A, Tetra-B, respectively. Tetra-C and Tetra-D, the tetrahedral DNA nanostructure probes are formed by self-assembly of the four ssDNA single strands. The specific information is: Tetra-A (75bp, molecular weight 23028.0, ssDNA), Tetra-B (55bp, molecular weight 17018.0, ssDNA modified with thiol at the 5' end), Tetra-C (55bp, molecular weight 16898.0, ssDNA modified with thiol at the 5' end ) and Tetra-D (55bp, molecular weight 16877.0, ssDNA modified with thiol at the 5' end), the four ssDNA single strands were purchased from Dalian Takara Bioengineering Co., Ltd. The four DNAs that constitute the tetrahedral structure probe contain three structural domains, each of which is complementary to the corresponding domains of the other three single-stranded DNAs (17 pairs of base...
Embodiment 2
[0076] Embodiment 2 sensitivity test
[0077] Different concentrations of hsa-miR-141 (1 aM, 10 aM, 100 aM, 1 fM, 10 fM, 100 fM, 1 pM, 10 pM, 100 pM, 1 nM, 10 nM) were detected.
[0078] Other steps of detection are the same as in Example 1, and the results are shown in Figure 3(A) and Figure 3(B), as can be seen from the results in the figure, the electrochemical signal increases with the concentration of target miRNA hsa-miR-141 (~10aM ) increases monotonously and changes linearly in the range from 1aM to 10pM. Using this curve, quantitative analysis of the target miRNA can be achieved. The results also show that when detecting hsa-miR-141 as low as 1aM, the electrochemical signal can reach 78nA, which is still much higher (>3SD) than the background signal (SD represents the standard deviation), indicating that the detection limit of the present invention can be lower to 1aM.
Embodiment 3
[0079] Embodiment 3 specificity test
[0080] The helper-miR-141 chain among the example 1 is changed into helper-let-7a, helper-let-7d respectively, for let-7 family miRNAs (let-7a, let-7b, let-7c, let-7d , let-7e, let-7f, let-7g, let-7i, miR-98) were detected to verify the specificity of the present invention, and hsa-miR-141 was used as a negative control. The sequences are as follows:
[0081] helper-let-7a: 5'-AGG CAA GAT GAA CTA TAC AAC CTA CTA CCT CAT GCG ACC T-3' (SEQ ID NO.7);
[0082] helper-let-7d: 5'-AGG CAA GAT GAA CTA TGC AAC CTA CTA CCT CTT GCG ACC T-3' (SEQ ID NO.8);
[0083] The biotin-modified DNA signal probe is the same as in Example 1.
[0084] Target miRNAs:
[0085] hsa-let-7a: 5'-UGA GGU AGU AGU AGG UUG UAU AGU U-3' (SEQ ID NO.9);
[0086] hsa-let-7b: 5'-UGA GGU AGU AGG UUG UGU GGU U-3' (SEQ ID NO.10);
[0087] hsa-let-7c: 5'-UGA GGU AGU AGG UUG UAU GGU U-3' (SEQ ID NO.11);
[0088] hsa-let-7d: 5'-AGA GGU AGU AGU AGG UUG CAU AGU U-3' (SEQ ID NO.1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com