Method for carrying out rapid amplification of whole blood genome
A genomic and rapid technology, applied in the field of biomedical molecular analysis and research, can solve the problems of high detection and amplification costs, false positive test results, non-specific amplification, etc., and achieves the omission of blood nucleic acid purification process, simple operation, and low cost. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
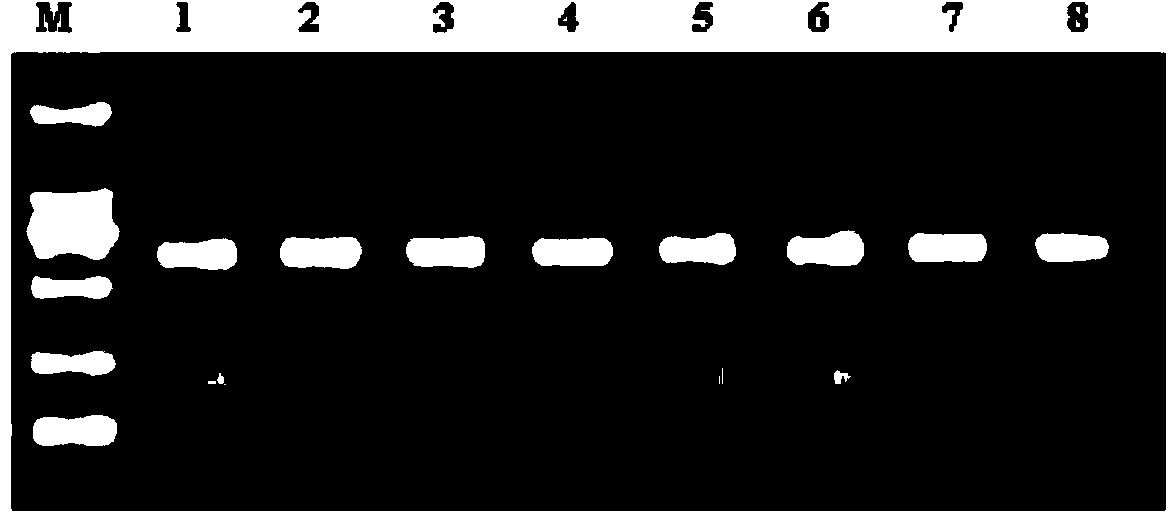
[0013] Use a pipette to draw 5 μl of nucleic acid release agent (0.25M NaOH, 20mM Chaps, 250mM (NH4) 2 SO 4 , 0.5M formamide) into a 0.2ml PCR reaction tube, then take 5μl of blood sample and briefly mix with it, and let stand at room temperature for 5 minutes. Then add PCR reaction solution (PCR reaction solution composition is 30mmol / L Tris-HCl (PH7.5), 10mmol / L (NH4) 2 SO 4 , 30mmol / LKCl, 4.5mmol / LMgCl 2 , 0.1-0.5M Betaine, 0.01-0.1%Gelatin, 0.5-5mg / ml BSA, 50-500mmol / L trehalose), 600μm / L dNTPs, 20pmol / L upstream and downstream primers, and 5U hot start Taq enzyme and 0.1U GP32 protein. Amplification was performed in a Bio-Rad PCR machine. After amplification, it was analyzed by 1.2% agarose gel electrophoresis.
[0014] The real-time PCR reaction system is 50 μl, and the PCR reaction program is shown in Table 1.
[0015] Table 1 PCR reaction program
[0016]
[0017] The experimental results are analyzed as follows:
[0018] (1) The sample has a wide range of ...
Embodiment 2
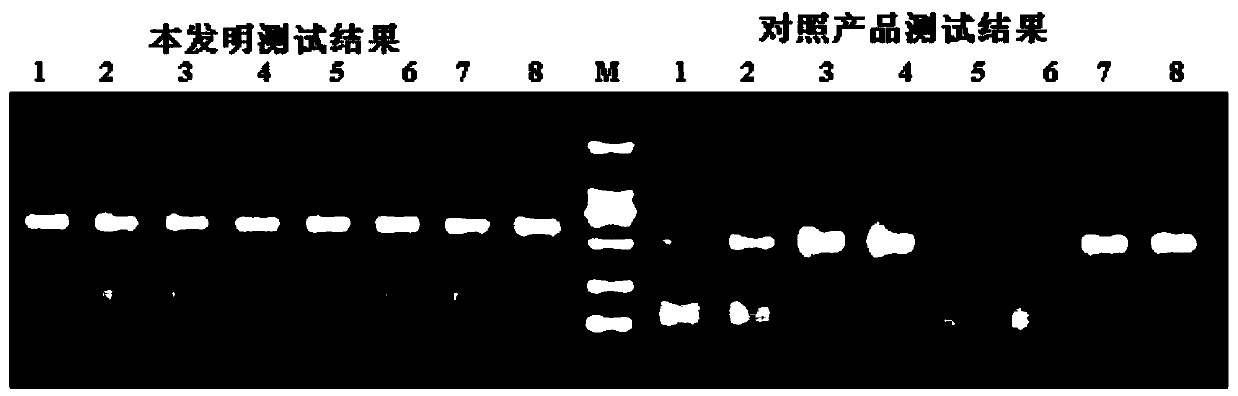
[0022] Compare with similar products in the market
[0023] The specific steps of the operation of the present invention are as follows: use a pipette to draw 5 μl of nucleic acid release agent (0.1M NaOH, 5mM Chaps, 750mM (NH4) 2 SO 4 , 0.1M formamide); add to a 0.2ml PCR reaction tube, then take 5μl blood sample and briefly mix with it, and let stand at room temperature for 5 minutes. Then add PCR reaction solution (PCR reaction solution composition is 30mmol / L Tris-HCl (PH7.5), 10mmol / L (NH4) 2 SO 4 , 30mmol / LKCl, 4.5mmol / LMgCl 2 , 0.1-0.5M Betaine, 0.01-0.1%Gelatin, 0.5-5mg / ml BSA, 50-500mmol / L trehalose), 600μm / L dNTPs, 20pmol / L upstream and downstream primers, and 5U hot start Taq enzyme and 0.1U GP32 protein. Amplification was performed in a Bio-Rad PCR machine. After amplification, it was analyzed by 1.2% agarose gel electrophoresis.
[0024] The specific operation process of similar products in the market is as follows: First, take 50 μl of PCR Mix reaction sol...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com