Real-time quantification PCR chip used for detecting gene expression of mouse cholesterol metabolism
A real-time quantitative and gene expression technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve problems such as lack of availability, no targeted research on genes, and limited accuracy of read numbers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
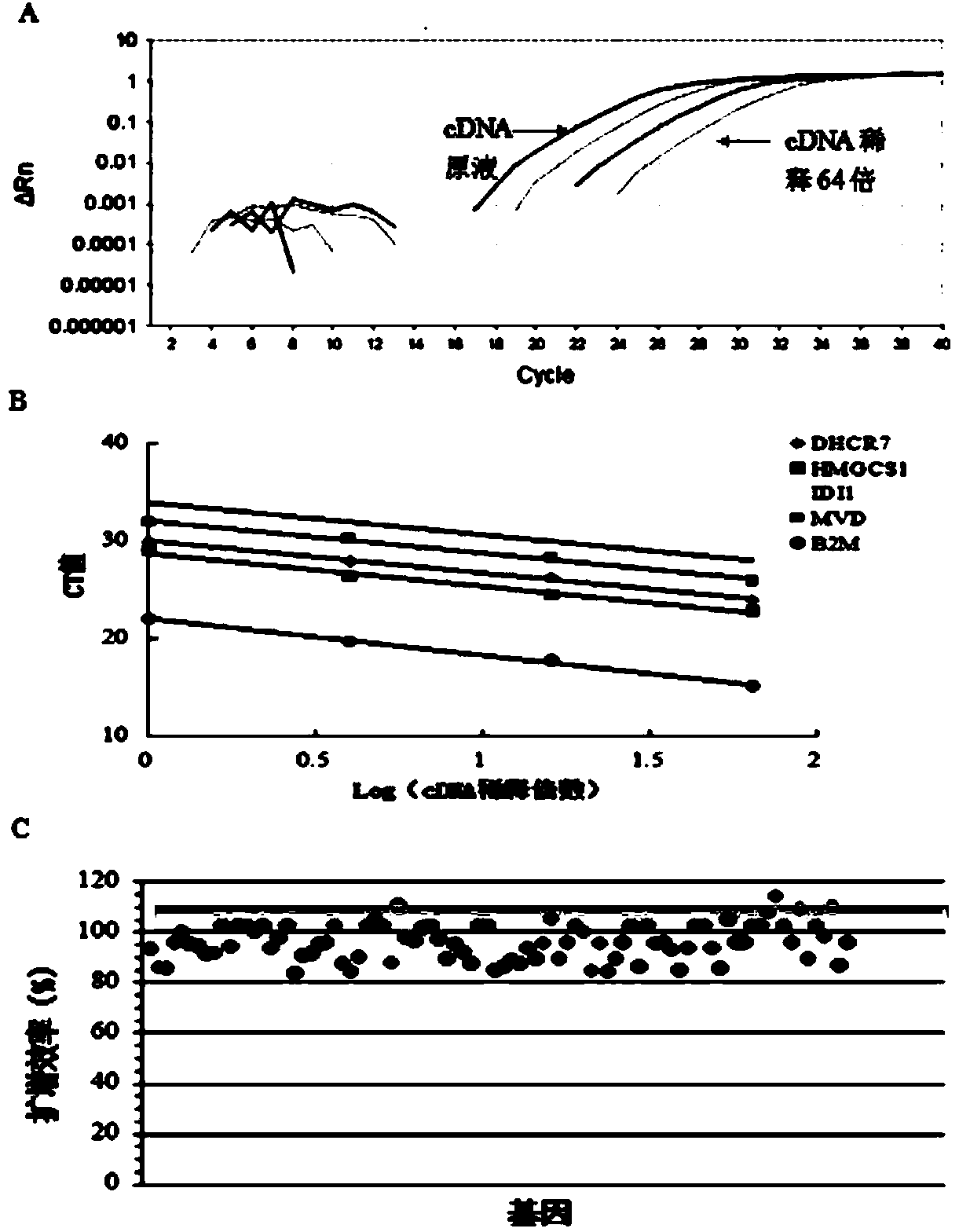
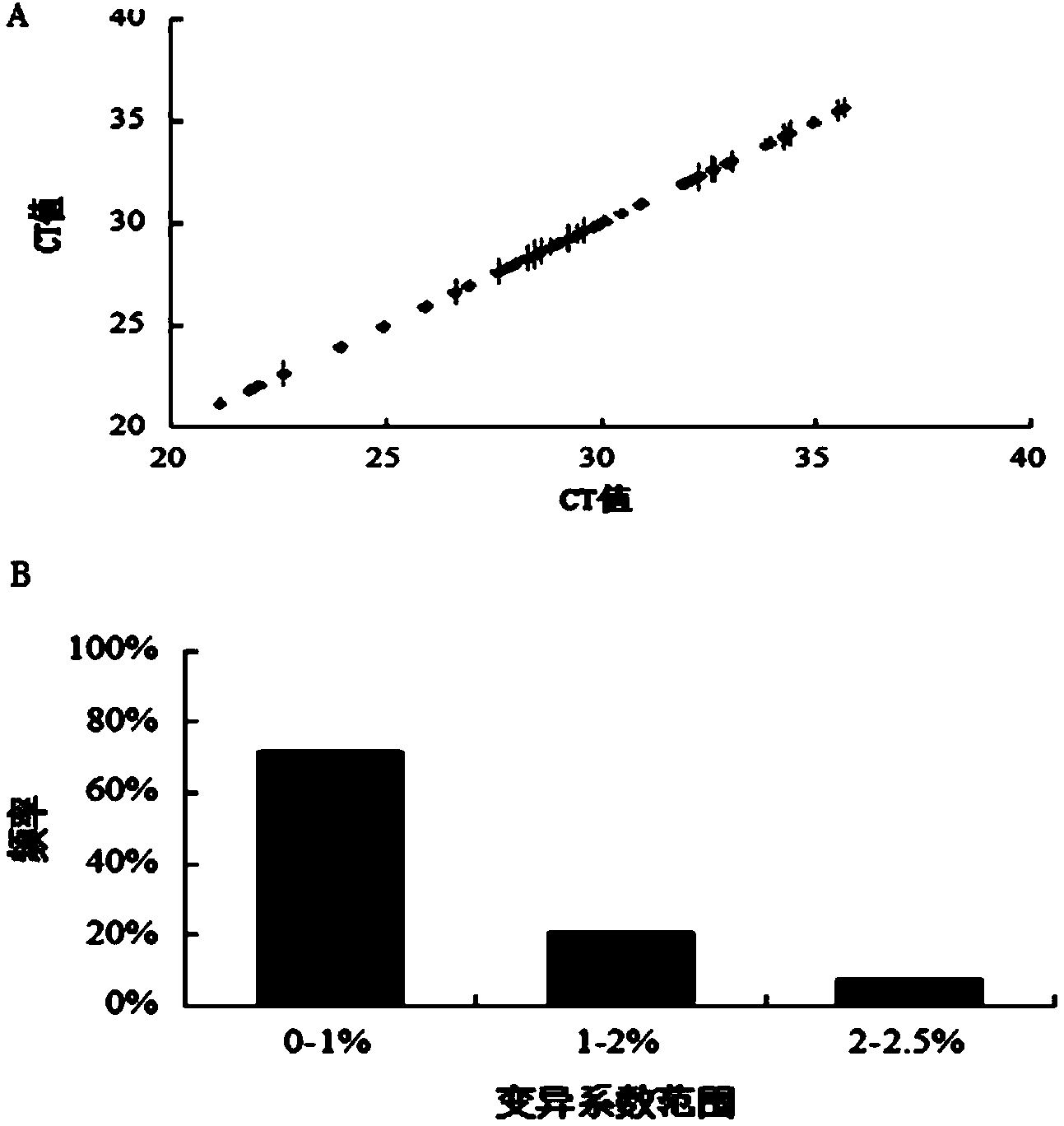
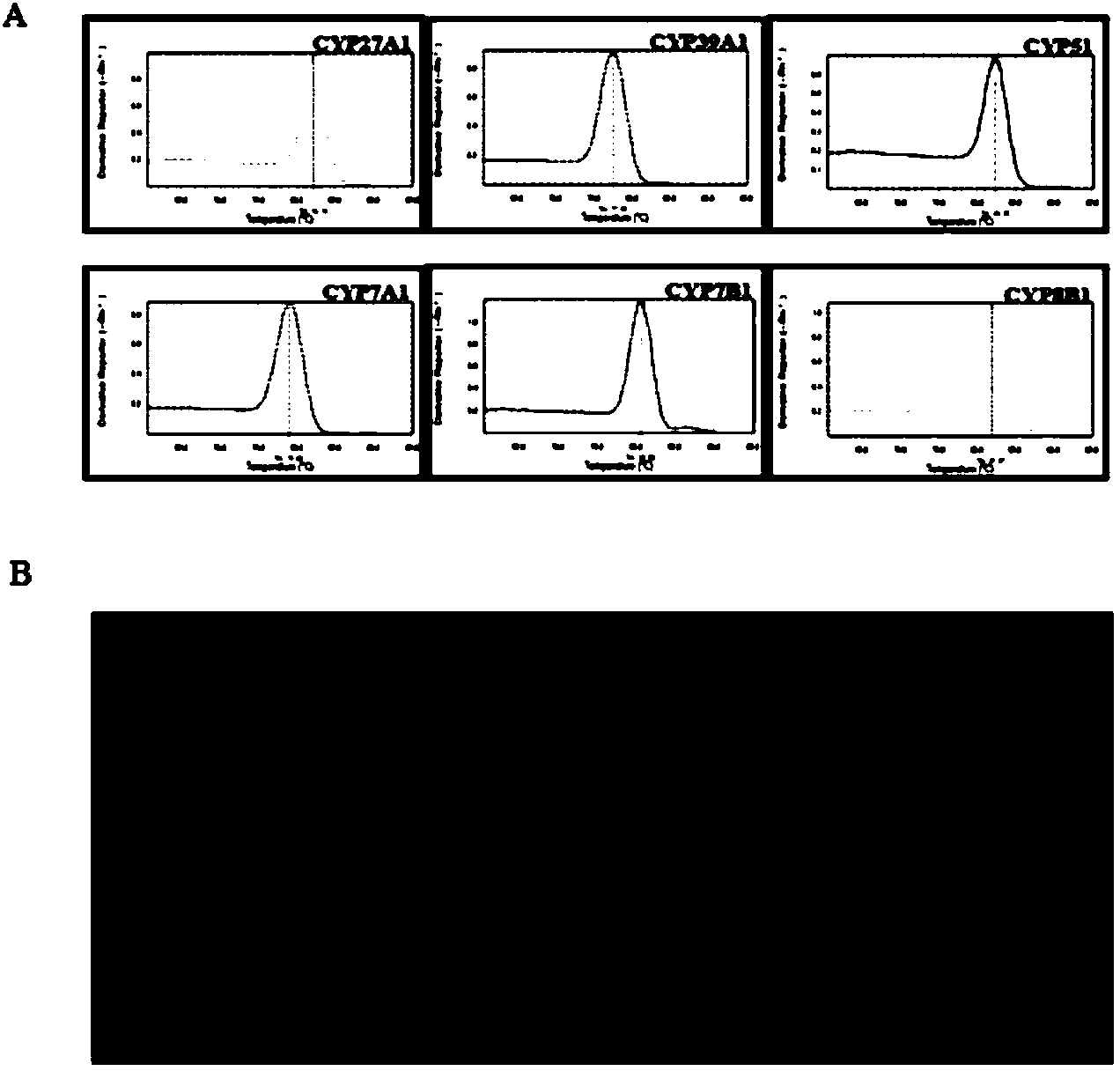
[0063] In this study, by designing a stable and reliable qPCR array detection scheme, mouse hepatocellular carcinoma cell Hepa1-6 was used as the research object, and qPCR array was used to detect the mRNA expression of cholesterol metabolism genes in Hepa1-6 induced by high glucose, aiming to explore the mRNA level. Effects of high glucose on genes involved in cholesterol synthesis.
[0064] 1 Materials and reagents
[0065] Mouse hepatoma cells Hepa1-6 were purchased from the Cell Resource Center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. Low DMEM medium (Hycolon, USA), fetal bovine serum (Hycolon, USA), D-glucose (Sigma, USA), Trizol (Invitrogen, USA), DNase1, reverse transcription reagent (Fermentas, USA), SYBR real-time PCR Premixture kit (Bioteke), PCR primers were designed and synthesized by Shanghai Sangon. Cell incubator (Thermo, USA), real-time fluorescent quantitative PCR instrument (ABI7500, USA).
[0066] 2 Cell culture, experime...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com