Recombinant protein of bay scallop peptidoglycan recognition protein as well as preparation and application thereof
A technology for recombinant protein and protein recognition, which is applied in the field of molecular biology and can solve the problems of insufficient research breadth and depth
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Example 1 The in vitro prokaryotic recombinant expression of the bay scallop AiPGRP gene coding region comprises the following steps:
[0021] 1. Construction of recombinant vector
[0022] The recombinant vector used in the present invention is the pEASY-E1 prokaryotic expression vector produced by Beijing Quanshijin Company. By PCR technique, using gene-specific primers P1 and P2 to amplify the AiPGRP gene of the bay scallop to remove the coding region fragment of the signal peptide. The reaction conditions were as follows: 94°C pre-denaturation for 5 minutes, followed by the following cycle: 94°C denaturation for 30 seconds, 60°C annealing for 30 seconds, 72°C extension for 45 seconds, a total of 35 cycles, and finally 72°C extension for 10 minutes. The PCR product was purified and recovered, and connected to the pEASY-E1 vector. After transformation, the colonies were screened by PCR and sequenced, and the plasmids of positive clones were extracted to complete the...
Embodiment 2
[0046] Example 2: Analysis of the coagulation activity of the bay scallop AiPGRP prokaryotic recombinant protein
[0047] Agglutination tests against Vibrio anguillarum, Micrococcus luteus and Pichia pastoris:
[0048] Vibrio anguillarum, Micrococcus luteus and Pichia pastoris cultured overnight were centrifuged, and the bacterial pellet at the bottom of the centrifuge tube was collected, and the collected pellet was washed with TBS (50mM L -1 Tris-HCl, 50mM L -1 NaCl) washed three times and then resuspended in FITC (final concentration 0.1mg mL -1 ) of 0.01M NaHCO 3 solution, and then marked in the dark for 30min. The labeled microorganisms were washed three times in TBS, and then resuspended in a solution containing 10mM ZnCl 2 in TBS buffer to make the concentration reach 2.5×10 9 cell mL -1 . Then take 10 μL of the above-mentioned microbial suspension and mix with 25 μL of the recombinant protein solution obtained in the above-mentioned examples (the concentration o...
Embodiment 3
[0050] Example 3: Antibacterial activity analysis of bay scallop AiPGRP prokaryotic recombinant protein
[0051] Growth inhibition experiments against Vibrio anguillarum and Micrococcus luteus:
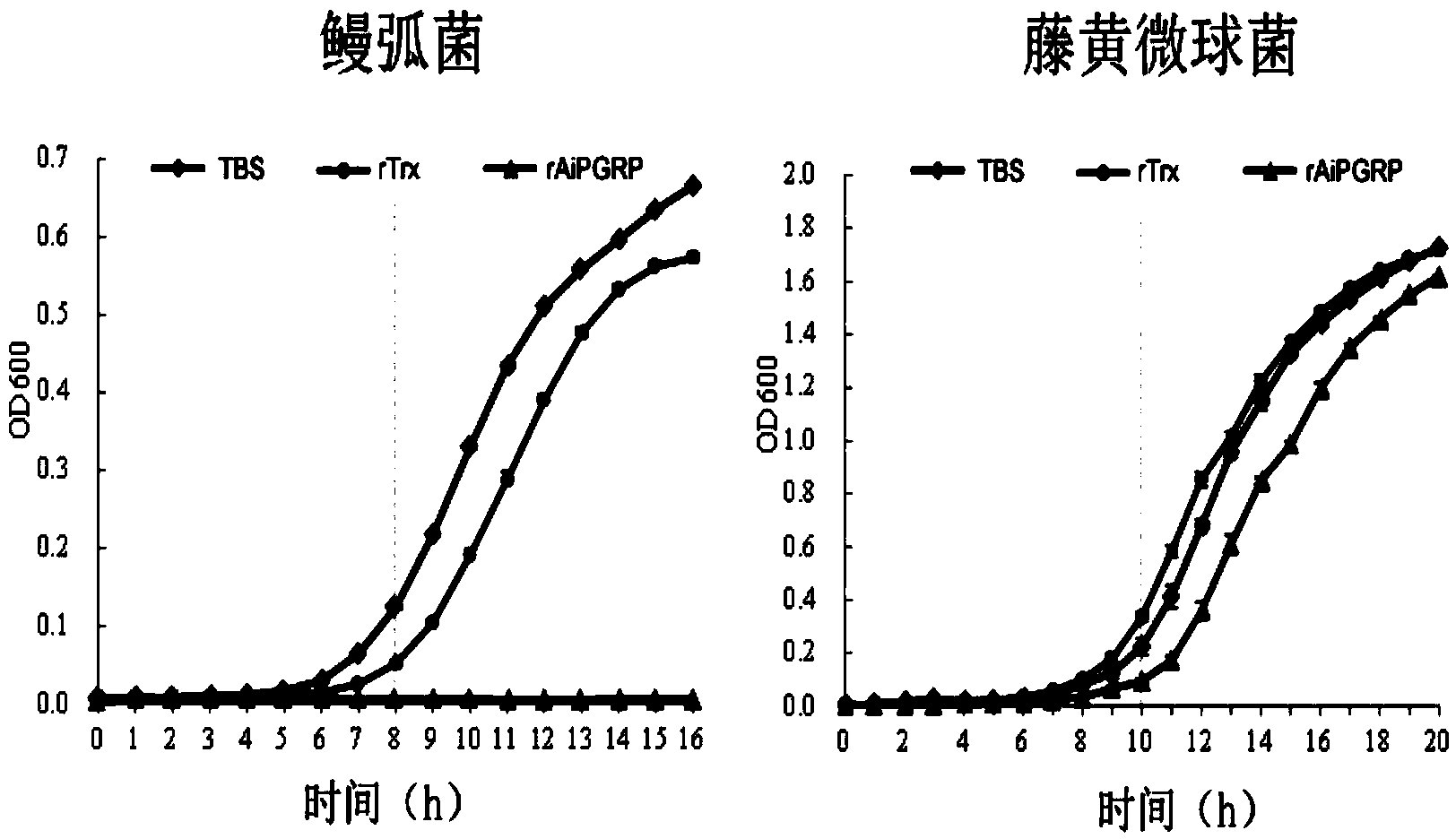
[0052] Add 200 μL LB liquid medium and 10 μL logarithmic growth phase of Vibrio anguillarum or Micrococcus luteus to each well of a flat-bottomed 96-well plate (Costar, Fisher), and then add the final concentration of 100 μg mL -1 The recombinant protein rAiPGRP, another rTrx protein control group and TBS blank control group were set up, all wells contained 10mM ZnCl 2 . Shake culture at 220 rpm at 37°C, detect and record the OD600 value of each well with a microplate reader every 1 h. Four repetitions were set up for each sample, and the average value was plotted according to the results of 4 measurements (see figure 2 ).
[0053] Experiments show that the recombinant AiPGRP protein can kill Gram-negative bacteria and inhibit the reproduction of Gram-positive bacteria.
[0054]...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com