Gene sequencing method
A gene sequencing, basic technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of poor DNA sequencing accuracy, complex devices, poor accuracy, etc., to improve sequencing accuracy and read Effects of long length, simple imaging device, and simplified synthesis steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
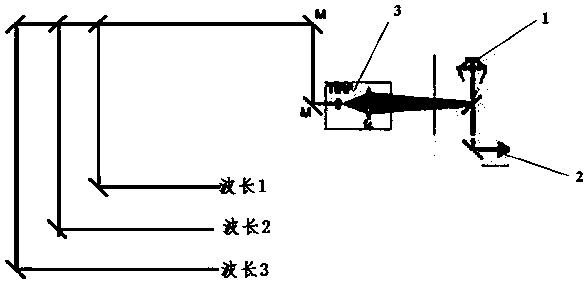
[0060] A gene sequencing method provided in this embodiment mainly uses lasers of three different wavelengths to excite three kinds of fluorescently labeled nucleotide molecules, and the nucleotide molecules are A, C, G and T. Any three of the nucleotide molecules, and the remaining one of the nucleotide molecules has no fluorescent nucleic acid label. This method redefines the sequencer with no fluorescence excitation as a symbol. It is a method that uses three-color fluorescence to synthesize and sequence.
[0061] Specifically, a method for gene sequencing provided in this embodiment includes the following steps:
[0062] (1) Synthesize a set of nucleic acid molecules for gene sequencing, including three fluorescently labeled nucleic acid molecules and one non-fluorescently labeled nucleic acid molecule, the three fluorescently labeled nucleic acid molecules are A, C, G and T four nucleotides Any three of the molecules, and the remaining one is a nucleic acid molecule witho...
Embodiment 2
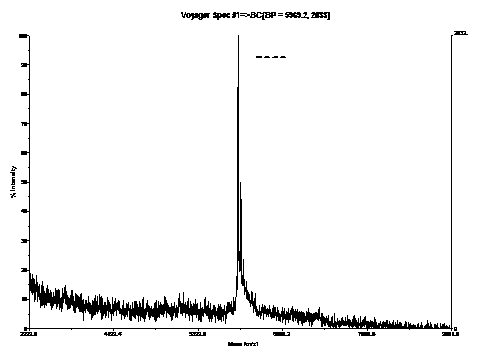
[0096] (1) Use the above-mentioned fluorescently labeled nucleic acid molecules to measure SNP (Single Nucleotide Polymorphism), and use mass spectrometry to identify the results:
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com