Method for distinguishing gynogenetic ctenopharyngodon idellus from common ctenopharyngodon idellus
A technology for gynogenesis and grass carp is applied in the field of molecular markers for identifying gynogenesis grass carp and common grass carp, which can solve the problems of easy confusion, limited detection sites, indistinguishable morphology and physiology, etc. Clear, consistent results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] (1) Genomic DNA extraction
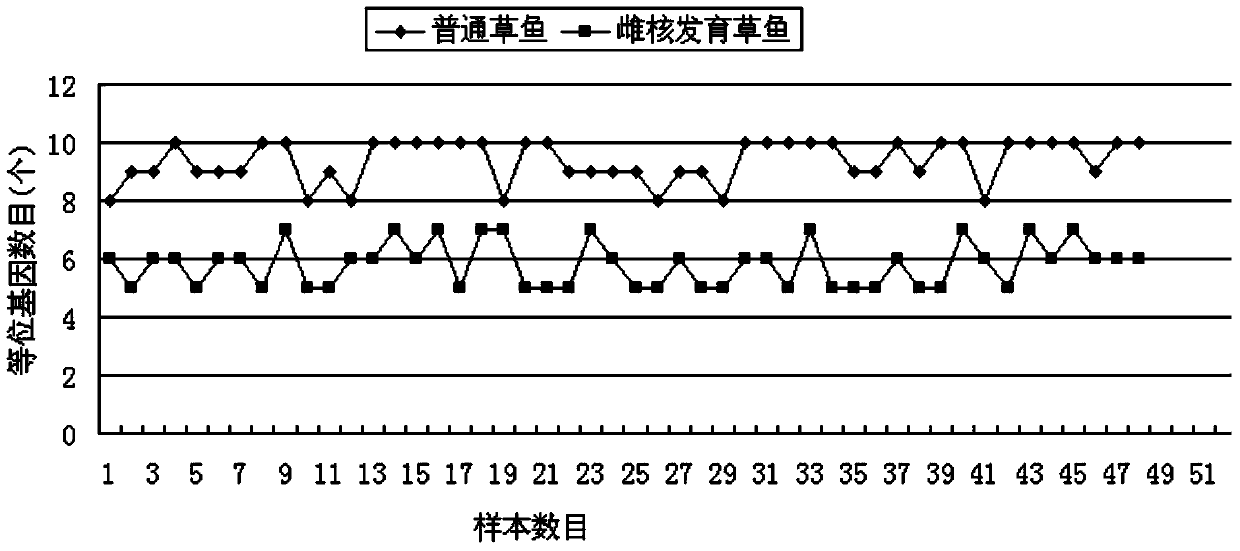
[0027] From May to July 2010, a group of 102 grass carp in the Yangtze River system were collected from three original breeding grounds of the four major domestic carps in Hubei and Hunan provinces to form the Yangtze River population. The sampling locations and numbers are shown in Table 1. Five Xingguo red carp males and 30 grass carp females from the Yangtze River system were used as parents, and the method of cold shock to inhibit the discharge of the second polar body was used to establish a heterospermous gynogenetic population of grass carp (approximately 100,000 fish in the level swimmer) . The gynogenetic grass carp progeny F1 population was used as the detection group (group G), and the wild grass carp population from the Yangtze River system was used as the control group (group W). 48 samples were randomly selected from each of the two groups, and the caudal fins were preserved in 95% Reserve in ethanol.
[0028] Table 1 Samplin...
Embodiment 2
[0052] (1) Genomic DNA extraction
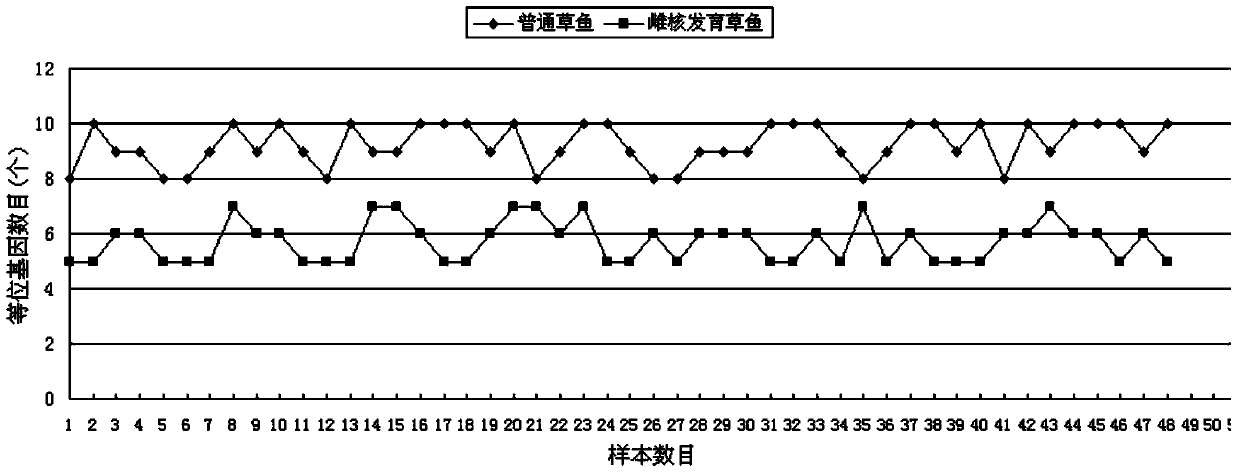
[0053] Similar to (1) in Example 1, the difference is that the method of inhibiting the first cleavage by heat shock is used to establish the gynogenetic population of grass carp.
[0054] (2) PCR amplification
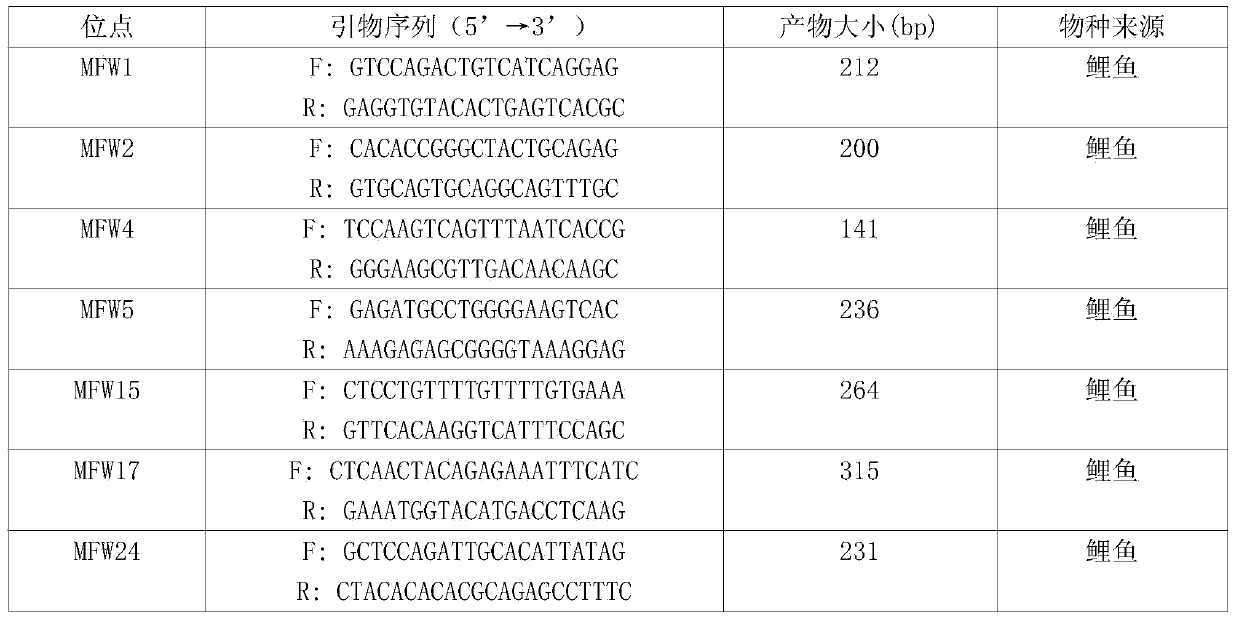
[0055] Using the genomic DNA solution prepared in step (1) as a template, 5 pairs of microsatellite primer pairs are used for PCR amplification respectively to obtain PCR amplification products. The SSR-PCR reaction system and procedure are as follows: 20 μL reaction system contains 10 μL of 10×buffer (reaction buffer), 0.5 μL of upstream and downstream primers, 40 ng of genomic DNA, and sterilized double-distilled water to make up the volume to 20 μL. The PCR amplification program was: 94°C pre-denaturation for 5 minutes, 94°C for 30s, 50-60°C for 30s, 72°C for 30s, 30 cycles, and 72°C for 7 minutes. The sequences of 5 pairs of microsatellite primers are as follows:
[0056] AAGGAACAGCATAAACCGAAAT GGAACCAAGCATCTGAAACTG
[0057]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com