PCR (polymerase chain reaction) product melting point analysis method for quantitatively detecting species composition of mixed-type sample
A quantitative detection and quantitative analysis technology, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, etc., can solve the problems of exacerbating negative effects, increasing specific primer interference, and difficult steps, so as to overcome errors and ensure specific detection , Create simple and fast effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
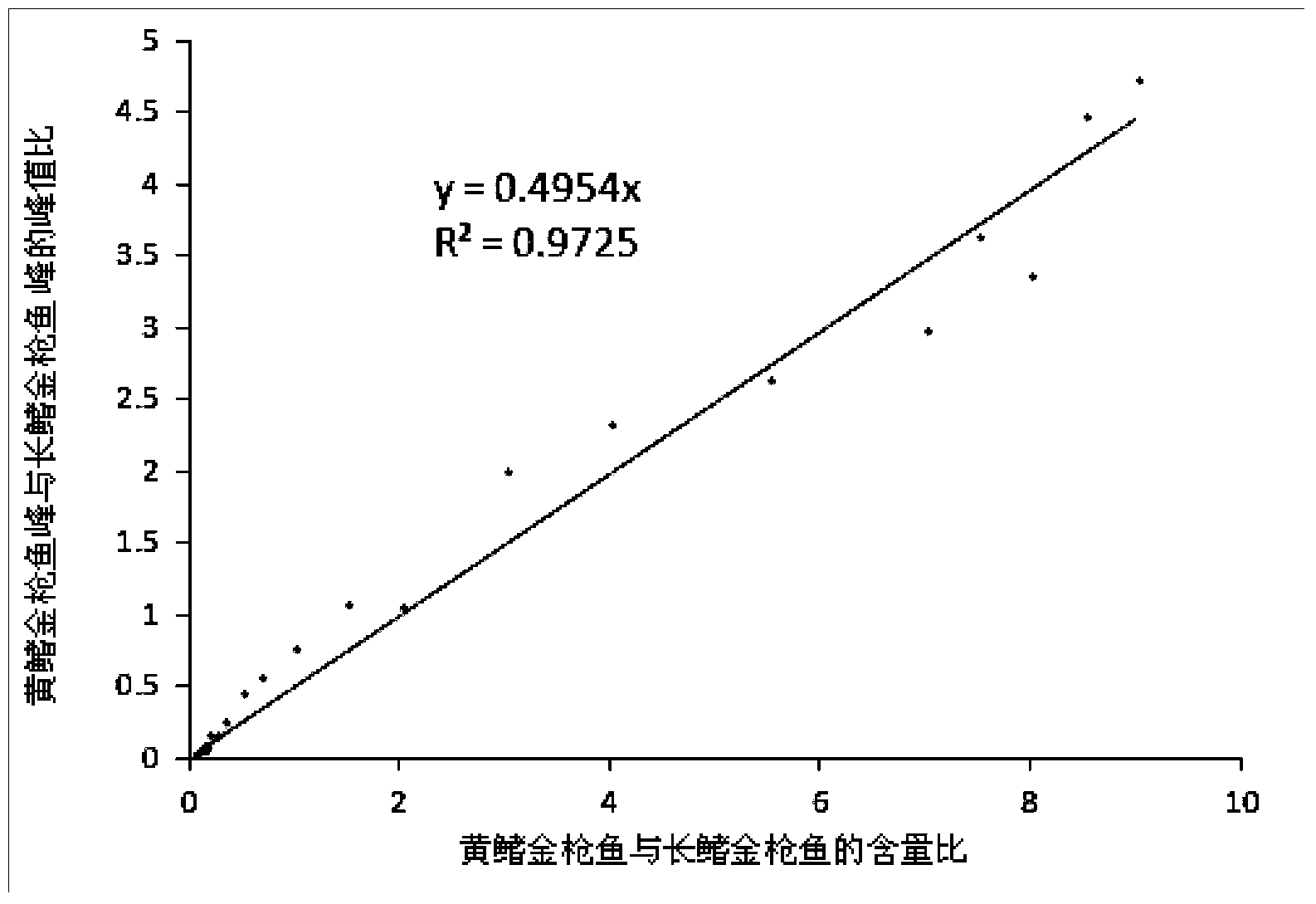
[0024] Embodiment 1 Albacore tuna and yellowfin tuna mixed sample
[0025] 1. Preparation of mixed samples with known component ratios and DNA extraction of mixed samples
[0026] The muscles of albacore tuna (Thunnus Alalunga, Albacore) and yellowfin tuna (Thunnus Albacares, Yellowfin tuna) were ground separately, mixed in a certain ratio and then ground to prepare a series of mixed samples with different mass ratios. DNA was extracted from the mixed sample, and DNA concentration and purity were determined using NanoDrop2000 (Thermo Scientific).
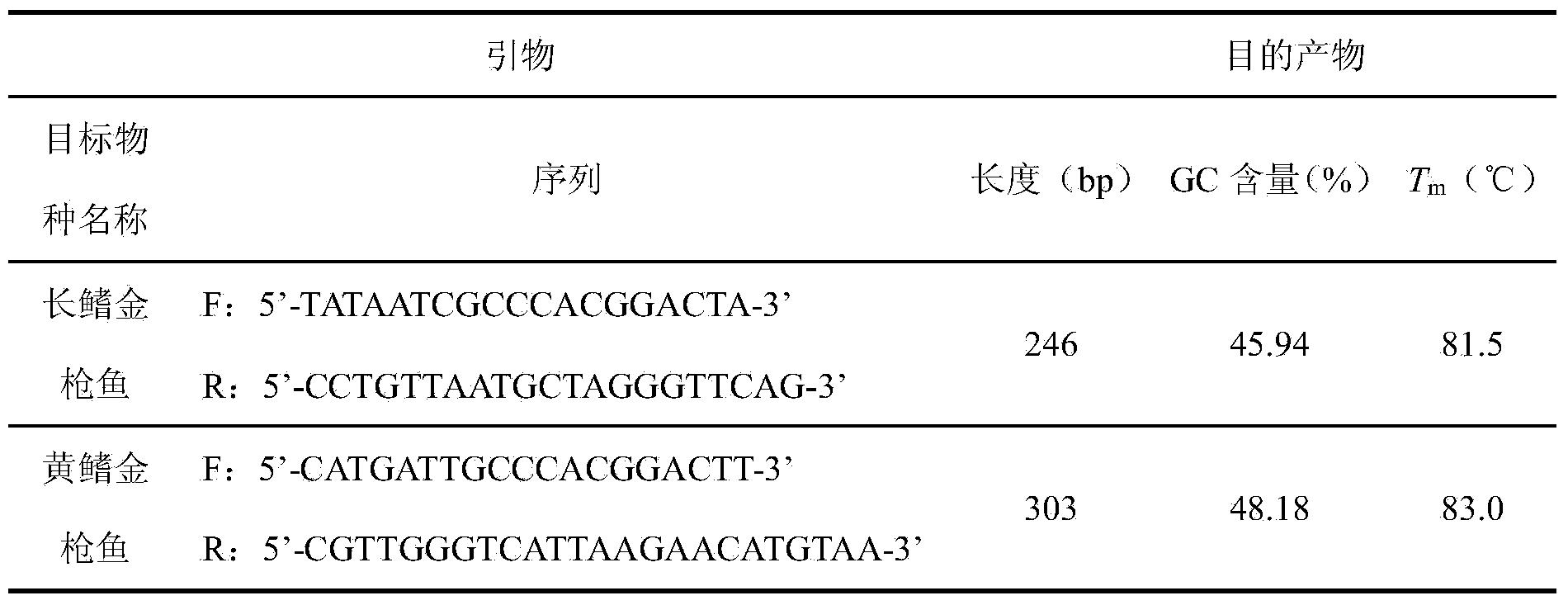
[0027] 2. Design of specific primers
[0028] Select the part of the ND4 gene encoding the hydrogenated coenzyme I dehydrogenase subunit in the mitochondria to design specific primers, and make the difference site at the key position of the primer extension, and use Primer Premier 6.0 software to match the primers. Performance is evaluated. Considering the GC content and length difference of the target fragment amplified by the p...
Embodiment 2
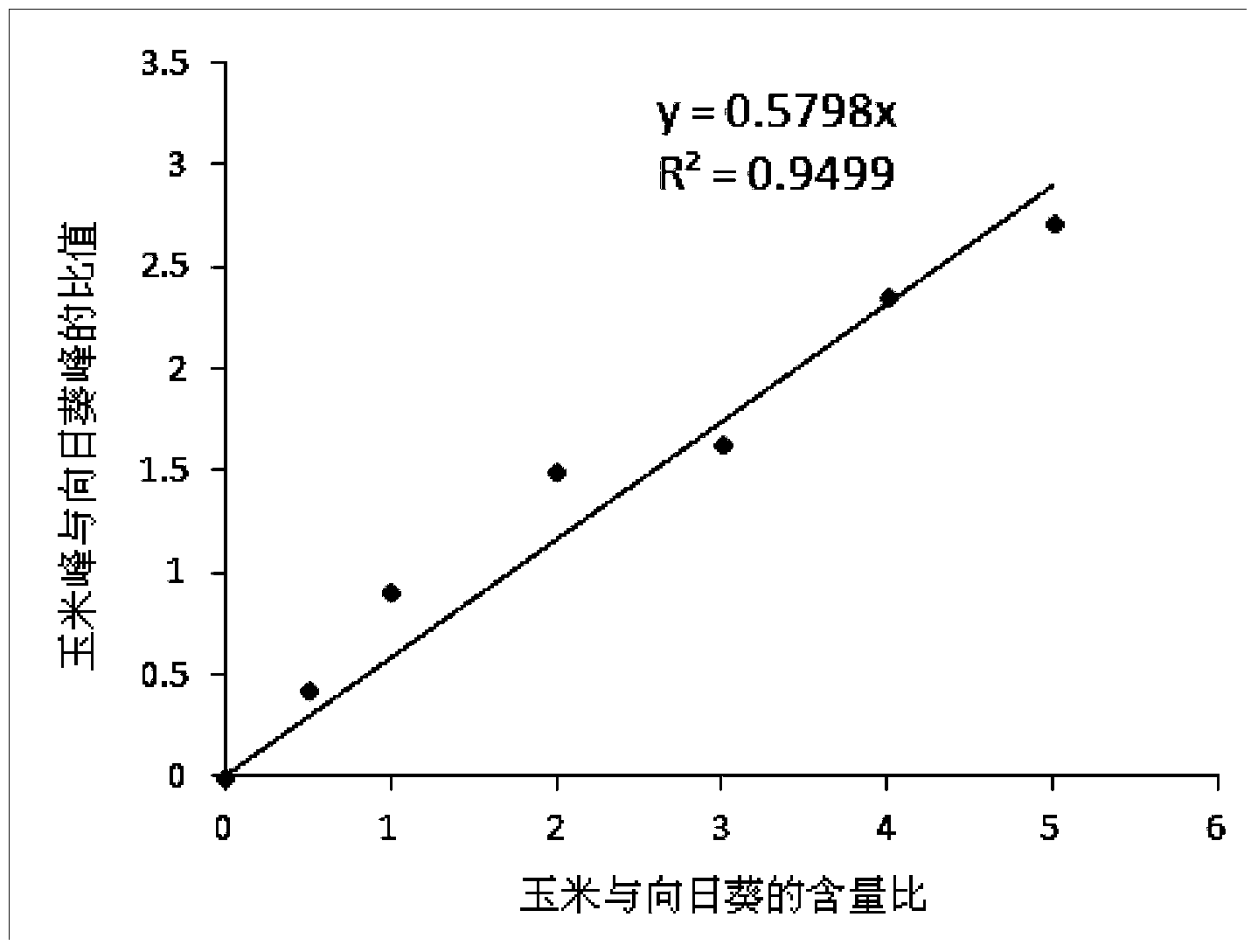
[0035] Example 2 Mixed samples of corn and sunflower
[0036] 1. Preparation of mixed samples with known component ratios and DNA extraction of mixed samples
[0037]Corn (Maize) and sunflower (Sunflower) germs were ground separately, mixed in a certain ratio and then ground to prepare a series of mixed samples with different mass ratios. DNA was extracted from the mixed sample, and DNA concentration and purity were determined using NanoDrop2000 (Thermo Scientific).
[0038] 2. Design of specific primers
[0039] The endogenous specific genes of maize and sunflower (maize zSSIIb gene, sunflower Helianthinin gene) were selected and specific primers were designed using Primer Premier6.0 software. And fully consider the difference in GC content and length of the target fragment obtained by primer amplification, and use ENDMEMO DNA T m Calculator estimates two target products T m Whether the value differs by more than 1.2°C. See Table 2 for the primer sequences and the GC co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com