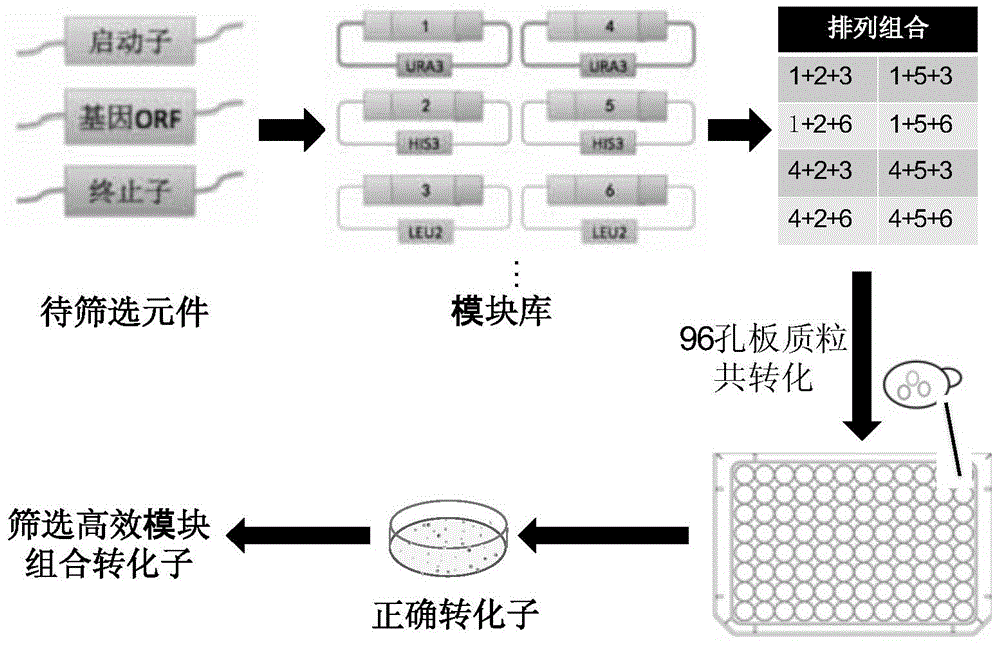
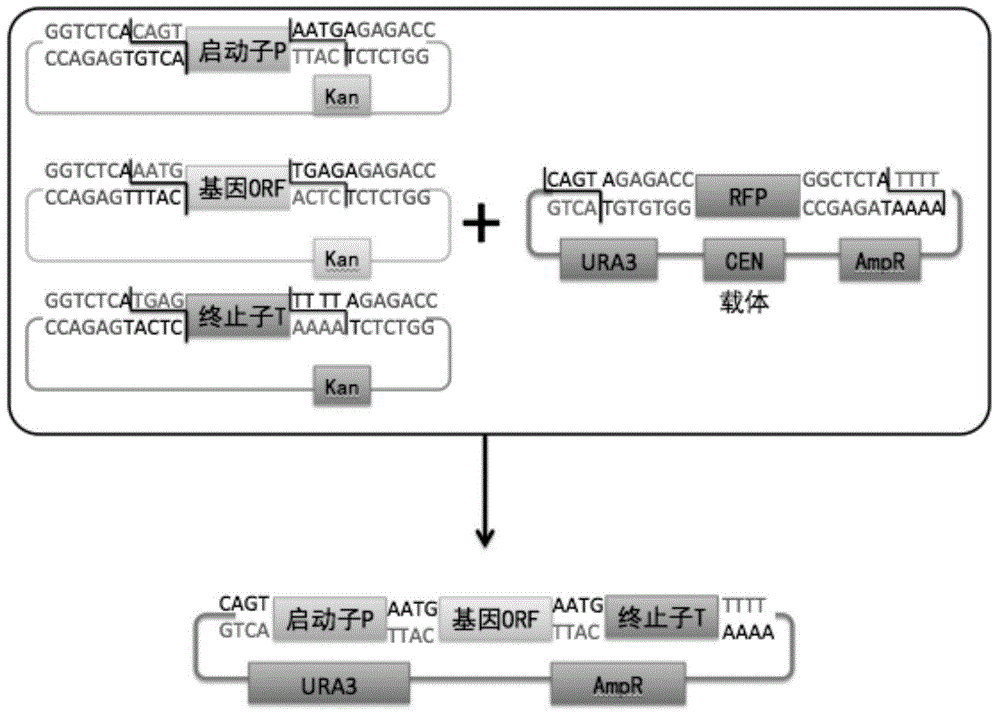
Saccharomyces cerevisiae module co-transformation combined screening method
A Saccharomyces cerevisiae module and screening method technology, which is applied in the field of Saccharomyces cerevisiae module co-transformation combination screening, can solve the problems of high price, one-sided research scope, combination type and number limitation, etc., to reduce workload, improve flexibility, and quickly screen Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] A combination screening method for Saccharomyces cerevisiae modules that efficiently utilize xylose
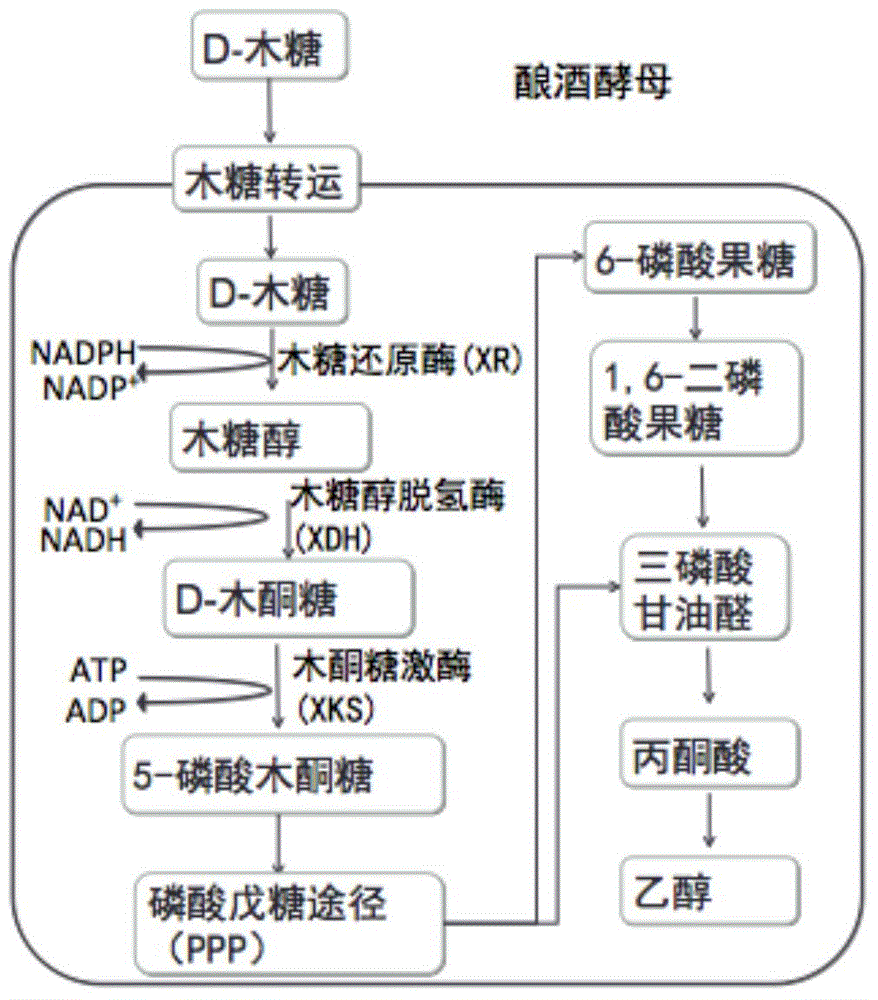
[0040] Saccharomyces cerevisiae is a traditional ethanol production strain with good industrial production characteristics. However, Saccharomyces cerevisiae cannot utilize xylose due to the lack of enzymes that convert xylose into xylulose in the xylose metabolic pathway, so it is necessary to introduce exogenous Pichia xylose reductase gene (XR), Hydrogenase gene (XDH) and overexpressed endogenous xylitol kinase gene (XKS), see figure 2 . Taking xylose reductase (XR) with different promoters, xylitol dehydrogenase (XDH) with different promoters and xylulokinase (XKS) with different promoters as an example, Screen and optimize the combination of promoters to obtain dominant strains with higher xylose utilization ability in xylose medium, which can be used for ethanol production.
[0041] (1) Each time select a promoter element, a gene element and a terminator eleme...
Embodiment 2
[0085] A Saccharomyces cerevisiae modular combination screening method for producing violacein metabolic precursor deoxychromoviridans
[0086] Violacein is a secondary metabolite synthesized from L-tryptophan as a precursor. It can be used as a potential antitumor, antiviral drug and biological dye, and has broad application prospects. Saccharomyces cerevisiae itself cannot produce the violacein metabolic precursors deoxychromoviridans and violacein. The target product of this example is the green violacein metabolic precursor deoxychromoviridans, and the exogenous genes that need to be introduced are vioA, vioB, and vioE (see Figure 8 ).
[0087] It is planned to introduce three exogenous genes vioA, vioB, and vioE into Saccharomyces cerevisiae, and fix vioE among them. By optimizing the combination of vioA and vioB gene promoters, it is expected to obtain the dominant combination of violacein metabolic precursor deoxychromoviridans Saccharomyces cerevisiae strain.
[008...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com