Method for constructing molecular map of bactrocera dorsalis intestinal bacteria colony
A technology of intestinal bacteria and Bactrocera dorsalis, applied in the field of bioengineering, can solve problems such as unclear microbial community structure and achieve important application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
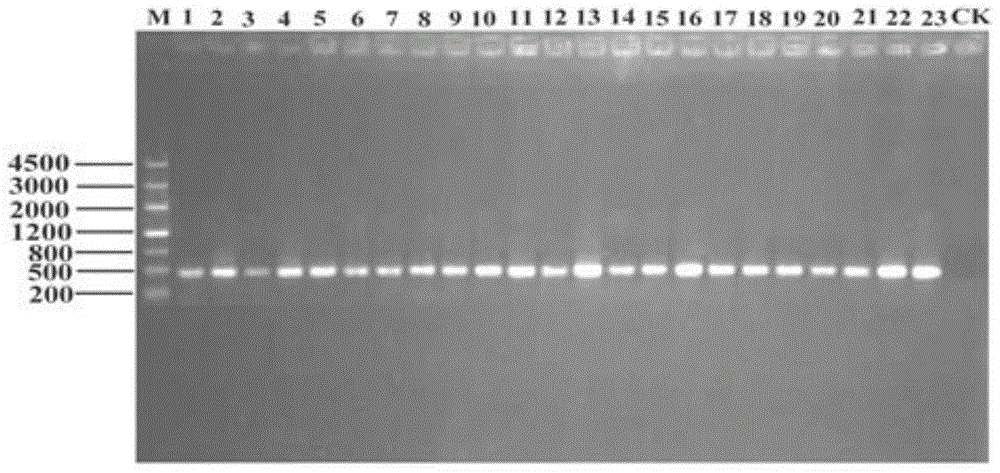
[0034] Example 1 Construction of the 16S rDNA library of Bacteralis dorsalis intestinal bacterial community
[0035] 1. Collection of samples
[0036] The adults used in the experiment came from three populations, namely the laboratory population (lab-reared (LR)), the laboratory sterile sugar-reared (LSSR) population and the wild population (field-collected (FC)) .
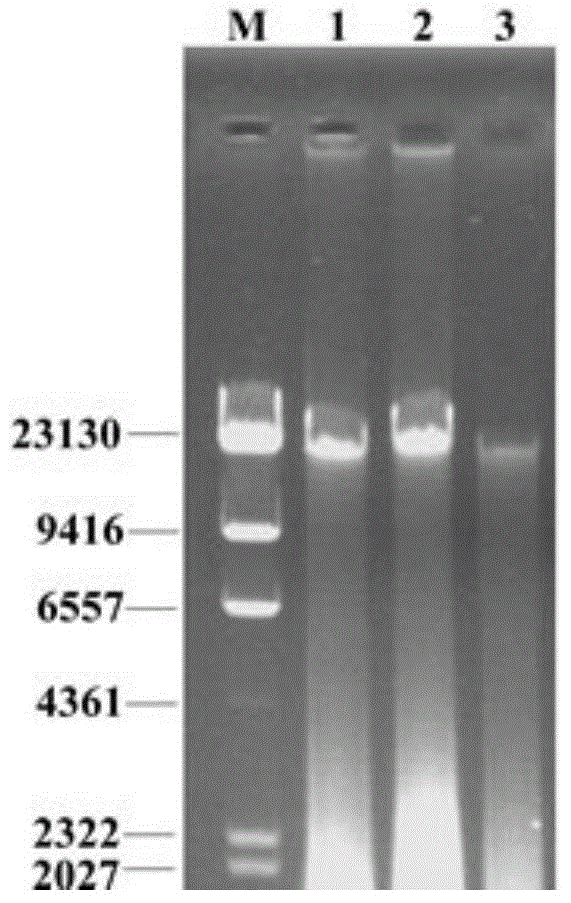
[0037] The laboratory population and the laboratory population fed with aseptic sugar water come from this laboratory, and the breeding conditions in the insectarium are: temperature 27°C±1°C, relative humidity 70%-80%, and the light cycle is natural light source. The laboratory population was fed with sucrose:yeast powder=1:3, and the sterile sugar water fed population was fed with 20% sucrose solution (sterilized by filtration). The wild population was collected from the orangery of the cadre rest center of the Bayi Road Military Region, Wuchang District, Wuhan, and brought back to the laboratory for intestinal...
Embodiment 2
[0060] Phylogenetic Analysis of Example 2 Bacterial 16S rDNA Library
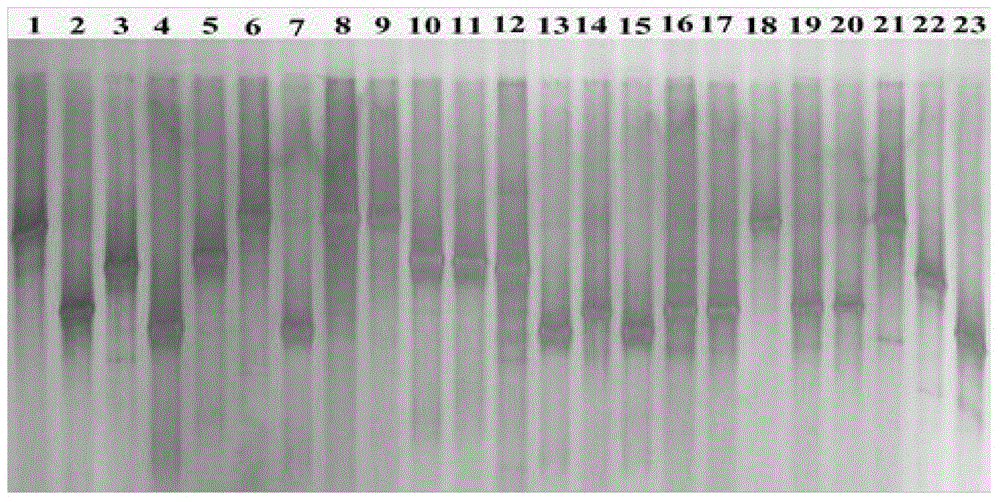
[0061] For the migratory position of the PCR amplification product obtained in Example 1 on the DGGE gel, select different recombinant plasmids (by visual observation, the bands at the same migratory position are classified as the same recombinant plasmid, which is the same phylogenetic type (phylotypes) or operational taxonomic units (OTUs)).
[0062] According to the DGGE electrophoresis pattern, 55, 26 and 69 different recombinant plasmids were screened out from 178, 181 and 187 recombinant plasmids in the three populations, and their inserts were sequenced (entrusted to Beijing Aoke Biotechnology Co., Ltd.) . Submit each sequencing result to the GENBANK nucleic acid sequence database for comparison with known sequences, and construct a phylogenetic tree with MEGA4.1. The obtained sequence was compared with the 16S rDNA gene sequence in GenBank by the BLAST program, the genetic distance was calculated ...
Embodiment 3
[0067] Diversity analysis uses the diversity index formula and Analytic Rarefaction 1.3 software to calculate the diversity index and coverage of the three libraries:
[0068] The coverage (C) was used to evaluate the expression of the constructed 16S rDNA library on environmental bacterial diversity, and the formula was:
[0069] C=1-(n1 / N)×100%,
[0070] In the formula, N represents the total number of clones in the 16S rDNA library, and n1 represents the number of OTUs that appear only once in the library.
[0071] For the obtained library 16S rDNA sequences, Shannon-Weiner index and Simpson index were used to describe the diversity of the community. The formula for the Shannon-Wiener index is:
[0072] H=-∑(P i )(㏑P i )
[0073] In the formula, Pi is the ratio of the number of clones of each OTU in the library to the total number of clones in the library.
[0074] The formula for the Simpson index is:
[0075] D=1-∑(P i ) 2
[0076] The Chao-1 index was used to e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com