A method for rapid diagnosis of mulberry sclerotinia
A technology for rapid diagnosis and sclerotinia disease, applied in the field of plant disease control, to achieve the effects of convenient and quick use, avoiding economic losses, and accurate and reliable detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Extraction of test material DNA. The aforementioned test strains were inoculated onto the PDA medium pasted with cellophane, and Sclerotinia, Botrytis cinerea and Trichoderma were cultivated in the dark at 20°C for 2 days, and the pathogenic bacteria of sclerotinia mulberry were grown at 25 Cultivate in the dark for 15 days and collect mycelium. Using the CTAB method ((Liu Li, Zhang Yongjun, Xu Changzheng, Luo Feng. An improved CTAB method to extract polysaccharide-producing fungal DNA. China Biotechnology Journal, 2014, 34(5): 75-79) to extract fungi and mulberry and other plants Genomic DNA of tissues and soil samples refer to Woodhall et al. cepivorumin soil. European Journal of Plant Pathology, 2012, 134(3): 467-473), the concentration and quality of all extracted DNA samples were compared by comparing the band intensity of agarose gel electrophoresis and measuring the 260 nm / 280 nm absorbance ratio for detection. The detected DNA samples were stored at...
Embodiment 2
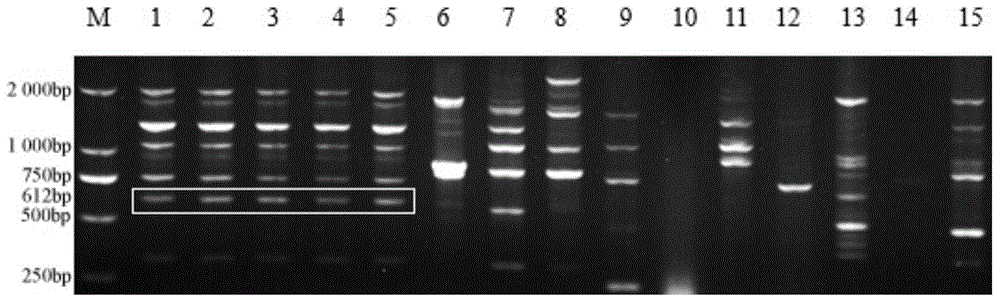
[0026] Example 2: Acquisition of specific conserved DNA fragments of G. nuclei. First, RAPD (random amplified polymorphism DNA)-PCR technology was used to amplify and screen specific conserved DNA fragment sequences of G. nuclei. The 40 random primers used were synthesized by Shanghai Sangong. Refer to the reaction procedure of Williams et al. (Williams J G K, Kubelik A R, Liv S ak KJ, Rafalski J A, Tingey V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 1990, 18(22): 6531-6535) , the total PCR volume of each RAPD amplification reaction is 25 μL, including 2.5 μL 10×PCR Buffer; 1.5 μL Mg 2+ (25mmol / L); 2μL dNTPs (2.5mmol / L); 1μL 10mM random primer, 1μL DNA template, 0.25μL rTaq enzyme (2.5U / μL); 2 O make up. The amplification program was as follows: pre-deformation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 36°C for 30 s, extension at 72°C for 1.5 min, 35 cycles; final extension at 72°C for 10 min ...
Embodiment 3
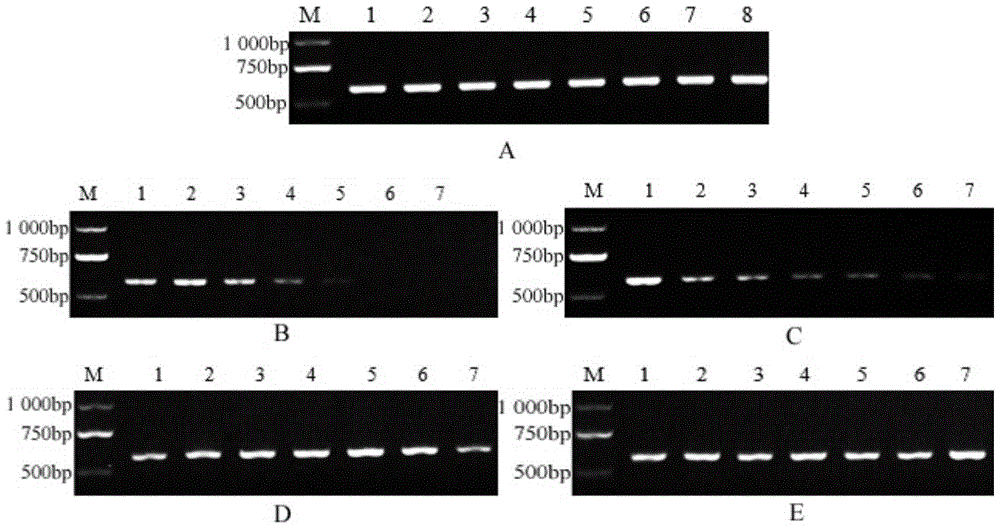
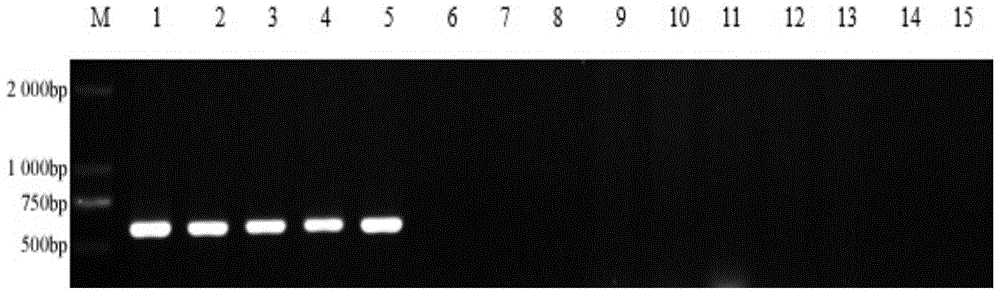
[0030] Example 3: Design and screening of SCAR-specific primers. According to the sequence of the specific fragment, 20 pairs of SCAR primers were designed with Premier 5.0 software, and the designed primer pairs were synthesized by Shanghai Sangon Biotechnology Co., Ltd. These 20 pairs of primers were screened by PCR, and a pair of best specific primers SS-F / SS-R were screened from these primers. The forward sequence SS-F of the primer is 5'- AGTAAAGAGAACATCAAACTTCG -3', located at 8 bp-30 bp of the specific sequence; the reverse sequence SS-R is 5'- ACTTCC ACCGACCA ATCT -3', located at 580bp-596bp of the specific sequence , the target fragment is 590bp (see the underlined part in the sequence listing).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com