Diabetes biomarkers and their applications
A technology of diabetes and microorganisms, applied in the field of biomedicine, can solve the problems that the pathogenesis of type II diabetes cannot be well explained, and type II diabetes needs to be improved.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1: Sample collection
[0055] All 344 stool samples were collected from 344 volunteers, and stool samples were collected by Peking University Hospital, Shenzhen, China. Diagnosis of type II diabetes was carried out according to the criteria issued by WHO in 1999 (Alberti, K.G. & Zimmet, P.Z. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabetic medicine: a journal of the British Diabetic Association 15,539-553, doi:10.1002 / (SICI)1096-9136(199807)15:73.0.CO;2-S(1998), which is incorporated by reference In this paper), the diagnosed type II diabetes patients were used as the case group, and other non-diabetic individuals were used as the control group (see Table 1). Type 2 diabetic patients and normal individuals need to provide frozen stool samples. Volunteers should pay attention to their diet 3 days before sampling. They ...
Embodiment 2
[0058] Example 2: DNA extraction and sequencing
[0059] 2.1 Storage of stool samples
[0060] Put the collected stool sample into the sterilized stool collection tube, and store it in the freezer immediately to freeze the stool sample. Frozen samples were sent to the storage point and stored at -80°C until use.
[0061] 2.2 DNA extraction
[0062]200 mg of frozen fecal samples were taken separately and suspended in a solution containing 250 μl of guanidine thiocyanate, 0.1 M Tris (pH 7.5) and 40 μl of 10% lauroyl sarcosine. The DNA extraction method was the same as above (Manichanh, C.et al.Reduced diversity of fecal microbiota in Crohn's disease revealed by a metagenomic approach.Gut 55,205-211, doi:gut.2005.073817[pii]10.1136 / gut.2005.073817 (by reference to 2006), incorporated into this article). DNA concentration and molecular weight were determined by Nanodrop instrument (ThermoScientific) and agarose gel electrophoresis, respectively.
[0063] 2.3 DNA library const...
Embodiment 3
[0067] Example 3: Identification of biomarkers
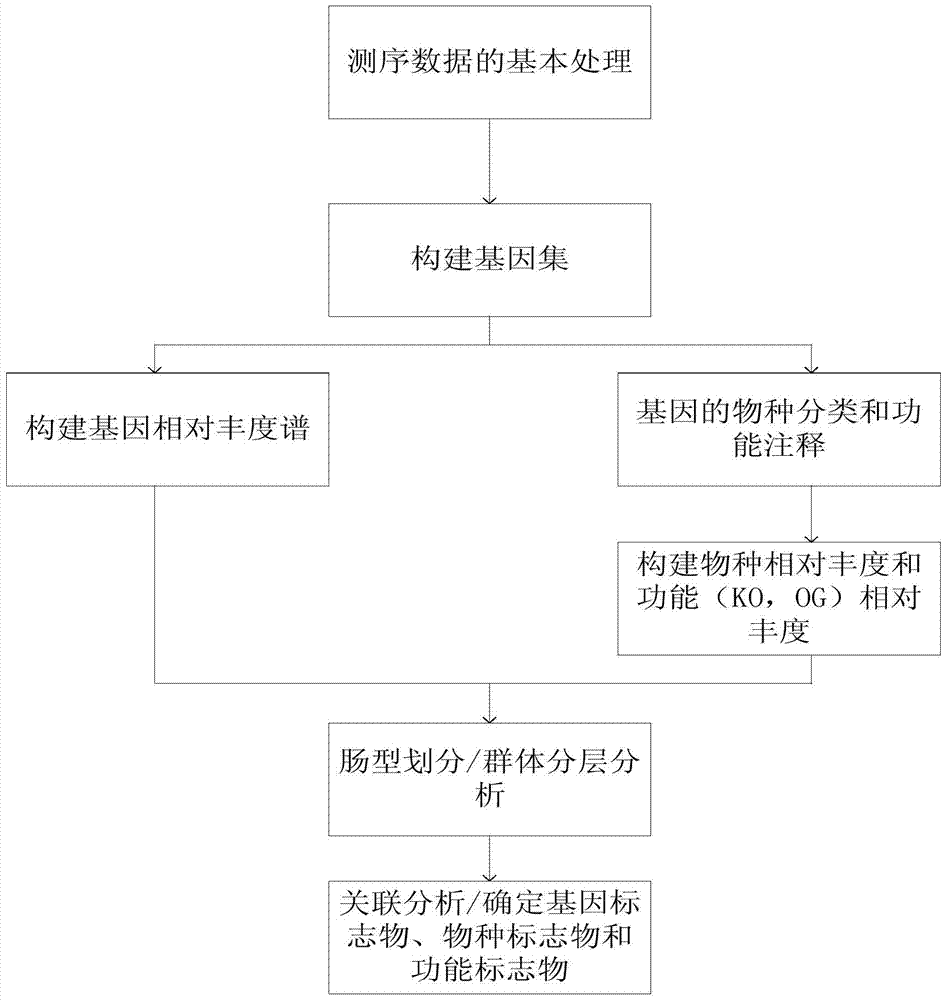
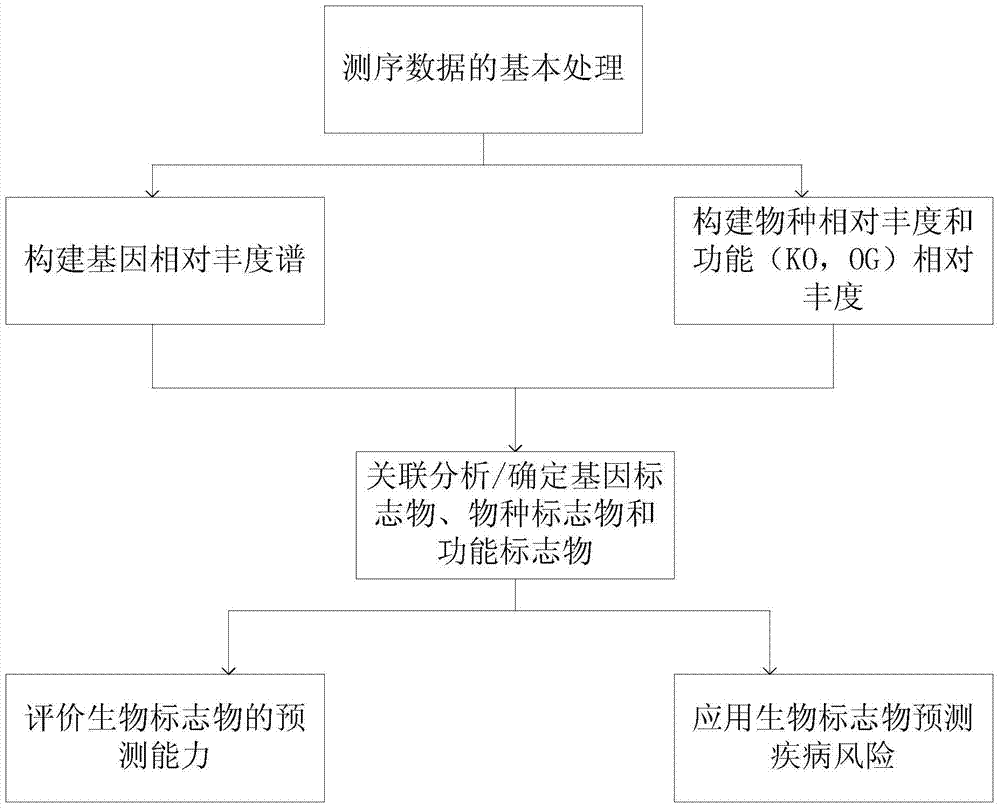
[0068] 3.1 Basic processing of sequencing data
[0069] After obtaining the sequencing data of the first phase of 145 samples, it was filtered to remove low-quality sequences containing 'N', adapter pollution sequences and host genome pollution sequences, and finally obtained 378.4Gb high-quality data. On average, high-quality data accounted for 98.1% of all data. In addition, the actual insert length of the PE library is between 313bp and 381bp.
[0070] 3.2 Updating the gene set
[0071] Using the same parameters as the MetaHIT gene set (Junjie Qin, Ruiqiang Li, Jeroen Raes, et al. (2010) A human gut microbial gene catalog established by metagenomicsequencing. Nature, 464:59-65, which is incorporated herein by reference), at In the first phase, use SOAPdenovov1.0642 and GeneMark v2.743 to perform de novo assembly and gene prediction on the sequencing sequence; then use BLAT software to compare all the predicted genes, if a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com