Method and kit for detecting deletion mutation of cell apoptosis regulator gene (BIM)
A detection method and gene deletion technology, which is applied in the fields of molecular biology and medicine, can solve the problems of time-consuming, high equipment requirements, and high false negative rate, and achieve the effects of reducing the possibility of pollution, simple method, and intuitive results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
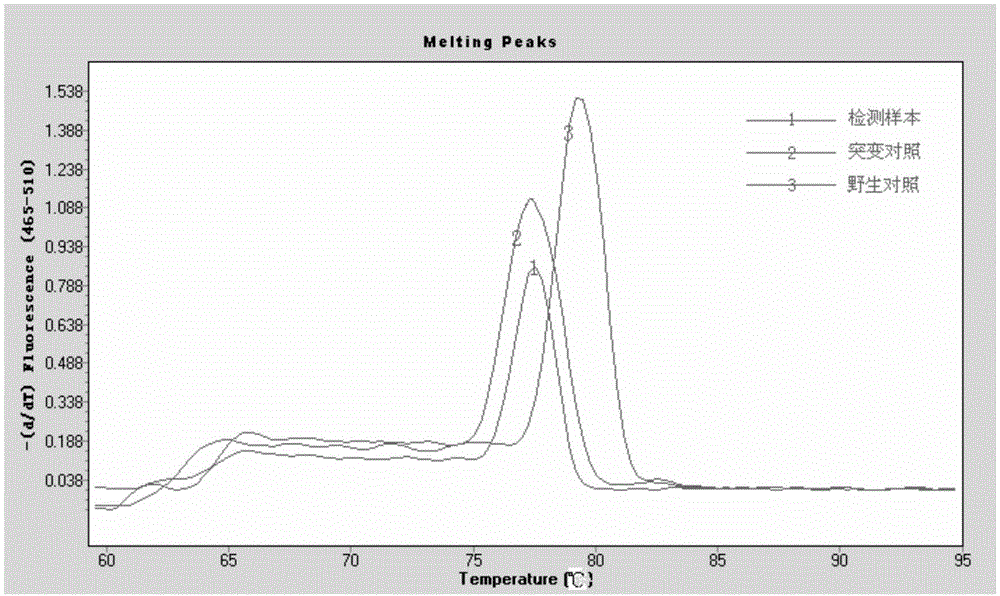
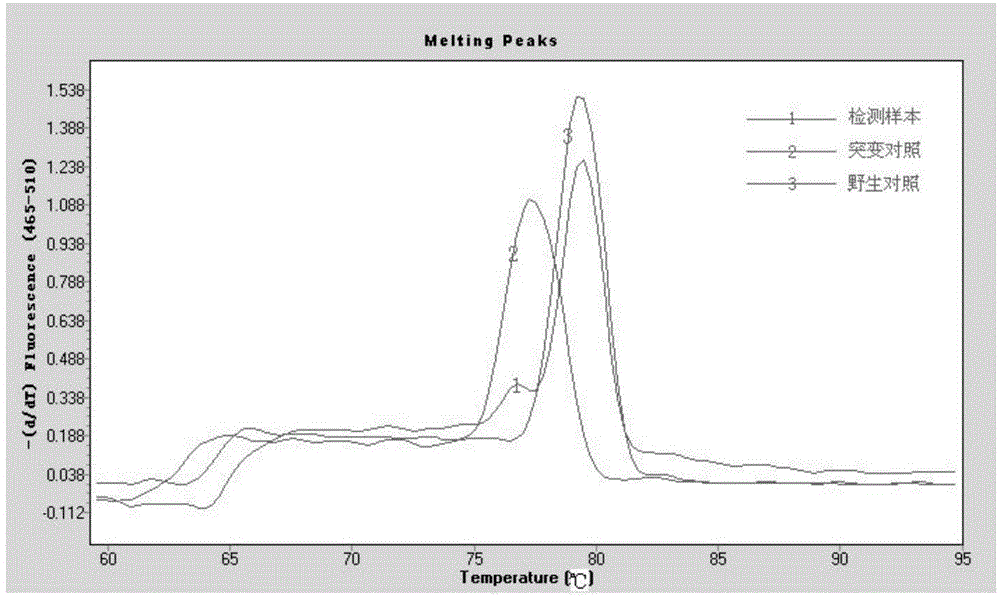
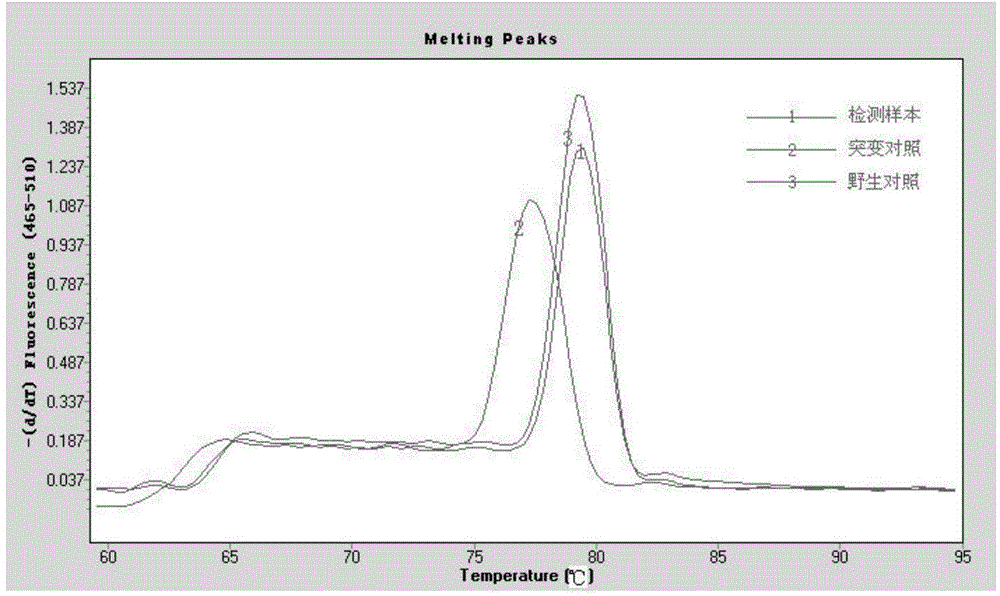
[0034] Embodiment 1 real-time fluorescent PCR detection
[0035] (1) Experimental materials
[0036] The 480II fluorescent quantitative PCR instrument was purchased from Roche Company of Switzerland, and the polymerase chain reaction solution (FastStart Universal SYBR Green Master) was purchased from Roche Company of Switzerland.
[0037] (2) Primer design and synthesis:
[0038] Using the partial sequence of the BIM gene as a template, the Primer Premier5 software was used to analyze and design primers, which were synthesized by Sangon Biotech.
[0039] Primers for detection:
[0040] BIM gene deletion detection upstream primer sequence:
[0041] 5'-ATACCATCCAGCTCTGTCTTCATAG-3' (SEQ ID NO 2)
[0042] BIM gene deletion detection downstream primer 1 sequence: 5'-CCCAACCTCTGACAAGTGACC' (SEQ ID NO 3).
[0043] BIM gene deletion detection downstream primer 2 sequence: 5'-TTGGTGGGAATGTAAAATGGC-3' (SEQ ID NO 4).
[0044] (3) Sample testing:
[0045] A total of 30 cases of l...
Embodiment 2
[0056] The 2903 bp deletion in the intron between the third and fourth exons of the BIM gene was detected by common PCR method, and 10 cases of the above-mentioned case samples and 10 control samples were selected for sequencing to determine the deletion of BIM.
[0057] (1) Experimental method
[0058] The PCR sequencing primers used above-mentioned fluorescent PCR primers were used, and the amplified products were subjected to agarose electrophoresis. The electrophoresis instrument was from BidRad Company of the United States.
[0059] (2) Experimental results
[0060] The screenshot of the result is as follows Figure 4 as shown,
[0061] In the end, the electrophoresis result of case X was the same as The analysis results of 480II fluorescent PCR were completely consistent.
[0062] (3) Association analysis of BIM gene 2903bp deletion and lung cancer susceptibility
[0063] The comparison of the distribution of BIM gene 2903bp deletion in lung cancer patients and con...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com