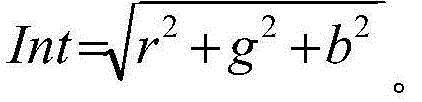Metalloprotease from exiguobacterium
A protease activity, protein technology, applied in the direction of enzymes, peptidases, hydrolases, etc., can solve the problem of limited use of metalloproteases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0352] Example 1. Sequencing of the deep-sea microbacterium genome
[0353] A novel bacterial strain was isolated from marine environmental samples collected in Denmark and designated as Exiguobacterium deep-sea, representing a new species within the genus Exigrebacterium.
[0354] Chromosomal DNA of this strain was isolated by QIAamp DNA Blood Mini Kit (QIAamp DNA Blood Mini Kit " (Qiagen, Hilden, Germany). 2 ug of chromosomal DNA was subjected to partial shotgun genome sequencing, a method in FASTERIS SA, Switzerland's commercially available service.By carrying out homology search with the known example of M4 metalloprotease, analyze the genome sequence of the protein sequence of coding this M4 metalloprotease and identify a kind of gene (SEQ ID NO: 1).
[0355] The nucleotide sequence and deduced amino acid sequence of the Exiguobacterium deep-sea gene are shown in SEQ ID NO:1. The coding sequence is 1536 bp including the stop codon. The encoded predicted protein is 511 ...
example 2
[0356] Example 2: Construction of the Bacillus subtilis expression strain comprising the deep-sea microbacterium genome sequence of the M4 metalloprotease polypeptide encoding SEQ ID NO:2
[0357] Cloning and expression of protease
[0358]As described in WO 99 / 43835 (hereby incorporated by reference), the signal peptide of the alkaline protease (aprH) from Bacillus clausii was fused in-frame to the DNA encoding the protease by SOE PCR fusion, thereby replacing Natural signal peptide. To amplify the coding DNA, the genomic DNA of Exiguobacterium was used as a template and the oligos SEQ ID NO:3 and SEQ ID NO:4 were used to amplify the gene by PCR, where the underlined sequence corresponds to the gene sequence And the sequence not underlined corresponds to the SOE expression cassette.
[0359] Forward primer GTTCATCGATCGCATCGGCT GAAGGTCTTCAATCTGGTAAG
[0360] (SEQ ID NO 3)
[0361] reverse primer GCGTTTTTTTATTGATTAACGCGT TTAGTAGACGCCAACTGC
[0362] (SEQ ID NO 4)
[...
example 3
[0384] Example 3: Purification of M4 Metalloprotease with SEQ ID NO:2
[0385] (The M4 protease was expressed in Bacillus subtilis.)
[0386] The broth was centrifuged (20000 x g, 20 min) and the supernatant was carefully decanted from the precipitate. The supernatant was filtered through a Nalgene 0.2 μm filter unit to remove remaining Bacillus host cells. The solid (NH 4 ) 2 SO 4 Added to the 0.2μm filtrate to a final concentration of 1.2M (NH 4 ) 2 SO 4 And the pH was adjusted to pH 6.9 with 3M Tris-base. The M4 protease solution was applied in 20mM HEPES / NaOH, 2mM CaCl 2 , 1.2M (NH 4 ) 2 SO 4 , on a Phenyl Sepharose FF (High Substitution) column (from GE Healthcare) equilibrated in pH 7. After washing the column well with equilibration buffer, M4 protease was treated with a linear gradient in equilibration buffer with 20mM HEPES / NaOH with 25% (v / v) 2-propanol, 2mM CaCl 2 , pH 7 was eluted over 3 column volumes. Fractions from this column were analyzed for pro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com