Recombinant human hepatitis B virus core protein fused protein
A hepatitis B virus and fusion protein technology, applied in the field of molecular biology, can solve the problems of inability to form core particles and maintain the normal function of HBc, etc., and achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Embodiment 1, HBV1.1c- and HBc expression vector construction
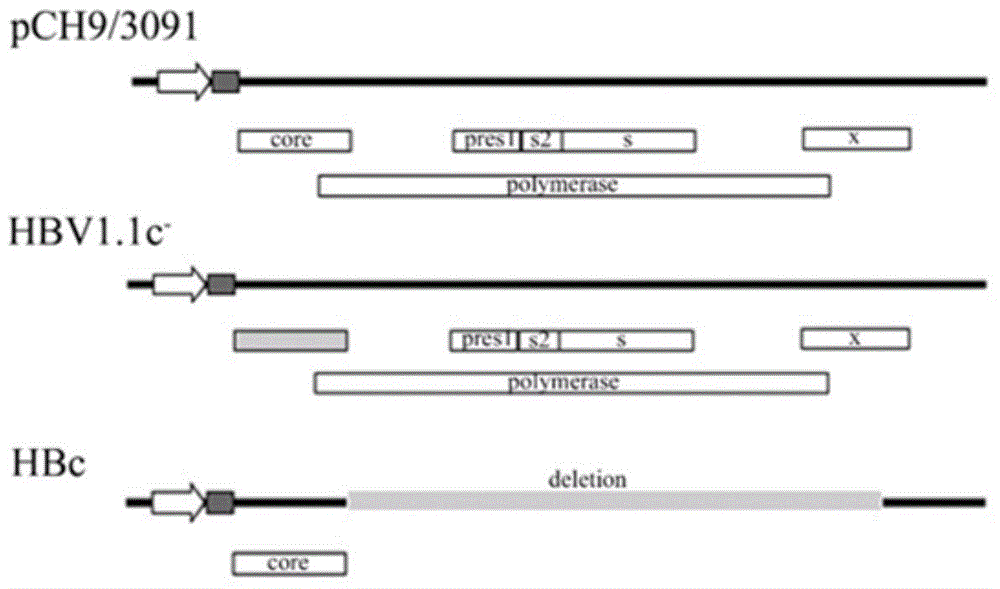
[0046] The successful replication of HBV DNA requires the joint action of core protein gene, HBV polymerase (Pol) gene and pregenomic RNA (pgRNA).
[0047] First construct the HBc plasmid containing HBV core protein gene as the basis of HBc transformation. The constructed HBc plasmid only expresses the HBc protein encoded by the Core gene in HBV, and does not express the Pol gene and pgRNA.
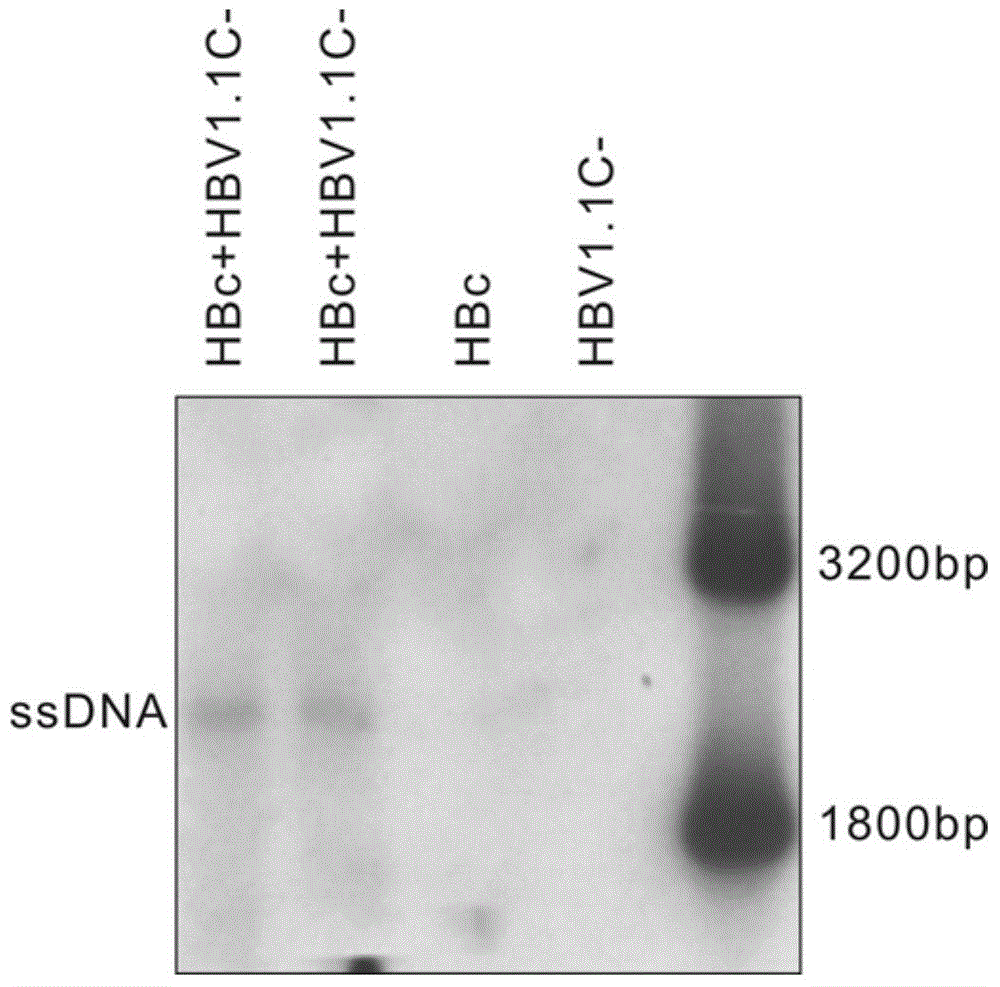
[0048]In order to test the function of the transformed HBc, it is also necessary to construct the plasmid HBV1.1c- which lacks the expression of HBc. Plasmid HBV1.1c- cannot express HBV core protein gene, but can express HBV polymerase (Pol) gene and pre-genomic RNA (pgRNA); HBc can provide HBV core protein in trans and support HBV DNA replication. HBV DNA replication can be restored when there are functionally normal HBc and HBV1.1c- present at the same time.
[0049] The method for constructing HBc is to remove mo...
Embodiment 2
[0065] Example 2: Construction of recombinant HBc with glycine-serine linkers of different lengths fused to the N-terminus
[0066] Recombinant HBc expression vectors with glycine-serine linkers with N-terminal fusion lengths of 47aa, 92aa, and 186aa were constructed.
[0067] 1. Construction of 12G4S-HBc
[0068] On the basis of the aforementioned HBc plasmid, construct 12G4S-HBc with 12 amino acid linkers fused to the N-terminus of HBc. The amino acid sequence of the 12G4S linker is: GSGGGGSGGGGS, and the nucleotide sequence of the 12G4S linker is: GGTTCAG GAGGTGGTGG ATCTGGAGGA GGTGGATCT. The specific construction method of 12G4S-HBC is:
[0069] (1) Use the upstream primer F12G4S: 5'-GCTTAGCCTTGGGTGGCTTTGGGGCATGGGTTCAGGAGGTGGTGGATCTGGAGGAGGTGGATCTGACATCGACCCTTAAAGA-3', and the downstream primer R1798: 5'-CCAATTTATGCCTACAGCCT-3' to amplify the HBc plasmid. Template, 0.5ul PrimeSTAR HS polymerase, 5×PrimeSTAR HS buffer 10μl, dNTP 4μl, and sterilized ultrapure water to make ...
Embodiment 3
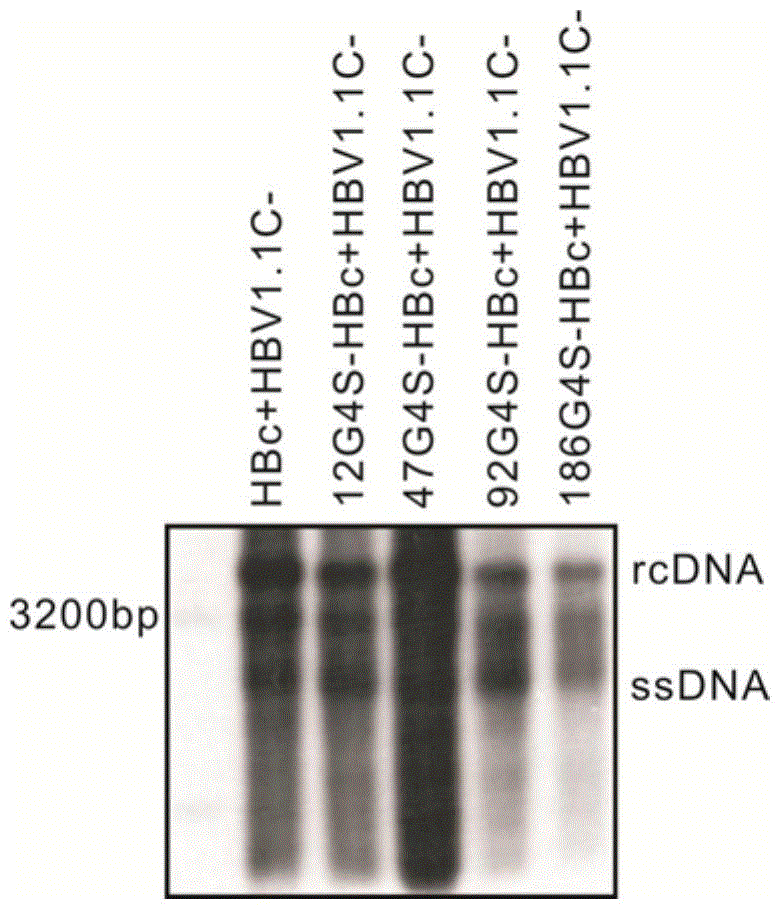
[0116] Example 3. Verification of the influence of the fusion of G4S linkers of different lengths on the normal function of HBc
[0117] The 12G4S-HBc, 47G4S-HBc, 92G4S-HBc and 186G4S-HBc constructed above were co-transfected with HBV1.1c- into HEK293 cells, and the cells were collected after 4 days, and the HBV DNA of the intracellular core particles was extracted, and then detected by Southern blot. The result is as image 3 As shown, when the above four kinds of glycine-serine fusion HBc proteins are co-transfected with HBV1.1c-, they can all support HBV replication, and it can be seen that the intermediate rcDNA and ssDNA are obviously replicated, and 47G4S-HBc even shows a significantly stronger replication level than HBc , while the HBV replication level of 92G4S-HBc and 186G4S-HBc is slightly lower than that of HBc, indicating that the insertion of long polyglycine sequences has less interference with HBc function, and even enhances HBc function in some cases.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com