Synechocystis-6803 genetically engineered bacterium capable of producing 3-hydroxypropionic acid, and construction method and application thereof
A technology of genetically engineered bacteria and trihydroxypropionic acid, applied in the direction of microorganism-based methods, biochemical equipment and methods, bacteria, etc., can solve the problem of complex separation and purification of cyanobacteria that have not yet been reported on genetically engineered bacteria that produce trihydroxypropionic acid , Large-scale industrial application gap and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Construction of Synechocystis 6803 Genetically Engineered Strain SMPA Producing Trihydroxypropionic Acid:
[0028] (1) In vitro amplification of the target gene:
[0029] Extraction of the genome of C. aurantiacus using bacterial genome extraction kit
[0030] Using SEQ ID No.1 and SEQ ID No.9 in the sequence list as the upstream and downstream primers, and the genome of C. aurantiacus as a template, the malonyl-CoA reductase gene mcr was amplified by PCR. The sequence of the gene mcr was used SEQ ID No. 17, shown as SEQ ID No. 2, SEQ ID No. 3, SEQ ID No. 4, SEQ ID No. 5, SEQ ID No. 6, SEQ ID No. 7, and SEQ ID No. 8. The sequences shown are the upstream primers, respectively, with SEQ ID No. 10, SEQ ID No. 11, SEQ ID No. 12, SEQ ID No. 13, SEQ ID No. 14, SEQ ID No. 15, and SEQ ID No. 16. The sequences shown are the downstream primers. The Bacterial Genome Extraction Kit was used to extract the Synechocystis 6803 genome, and the Synechocystis 6803 genome was used as a templat...
Embodiment 2
[0042] Conditions for 3-HP production in the constructed strain SMPA
[0043] The constructed strain SMPA was cultured in normal BG 11, and the setting parameters of the shaker were the light intensity of 2000 Lux, the rotating speed of 130 rpm, and the temperature of 30°C. Take OD during vaccination 730nm Add 5 mL of 0.2 fresh cells to 20 mL of culture medium. Make 3 parallel samples in each group. Use a UV-1750 spectrophotometer to measure the absorbance at a wavelength of 730 nm. 730nm When it is 1.0, centrifuge 25mL bacterial solution at 900xg for 15min, resuspend with 10mL fresh BG11 medium, cultivate and produce 3-HP, and add 1M NaHCO every day 3 0.5mL, the 3-HP production was measured after 6 days.
Embodiment 3
[0045] Expression and identification of malonyl-CoA reductase
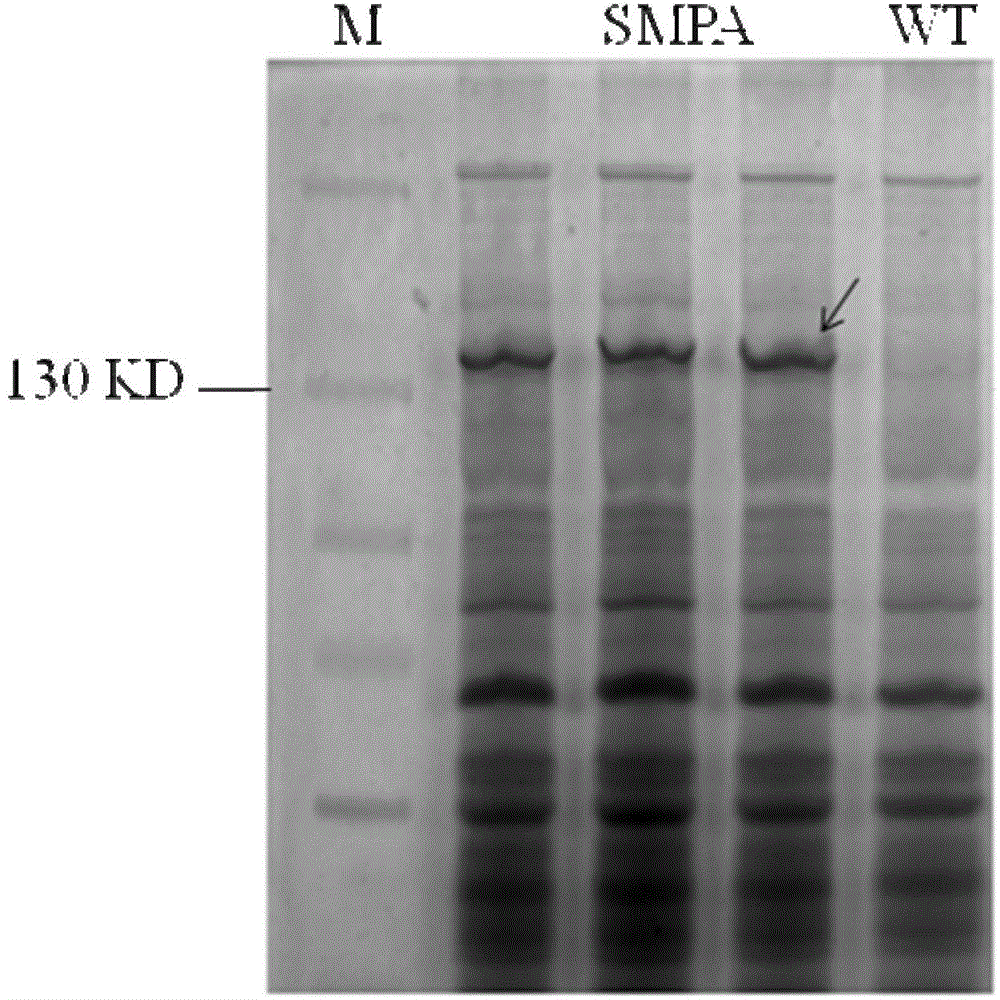
[0046] The cells were collected, centrifuged at 7600 rpm for 10 minutes, and the cells were kept in a boiling water bath at 100°C for 10 minutes, and then subjected to SDS-PAGE electrophoresis ( image 3 ).
[0047] From image 3 Comparing the total protein of the constructed strain and the wild-type strain, it can be observed that the malonyl-CoA reductase is expressed and its size is 132KD.
[0048] 3-HP production detection
[0049] Cells were collected, centrifuged at 7600rpm at 4°C for 10min, the supernatant was left for sample derivatization, 100μl of supernatant was taken, and after drying, 10μl of methoxyamino hydrochloride pyridine solution (4mg / ml) was added with 30°C 750rpm constant temperature shaking for 90min. Add 90μl MSTFA 37°C 450rpm constant temperature shaking for 30min. Centrifuge at 13000rpm for 10min at room temperature, take 70~75μl and load the sample, taking care not to absorb the white floccules...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com