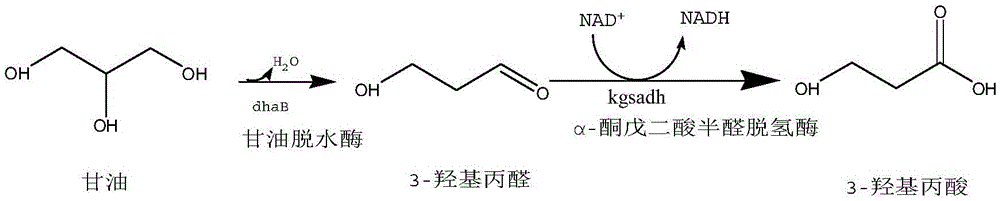
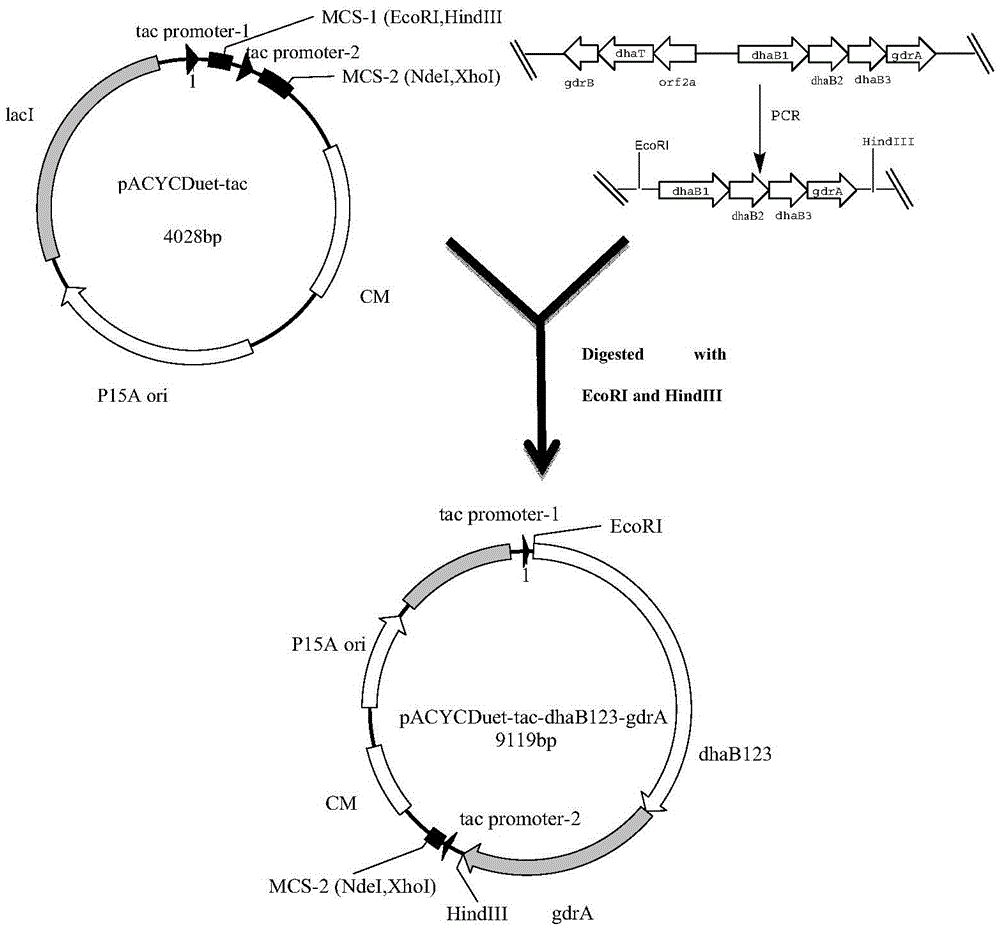
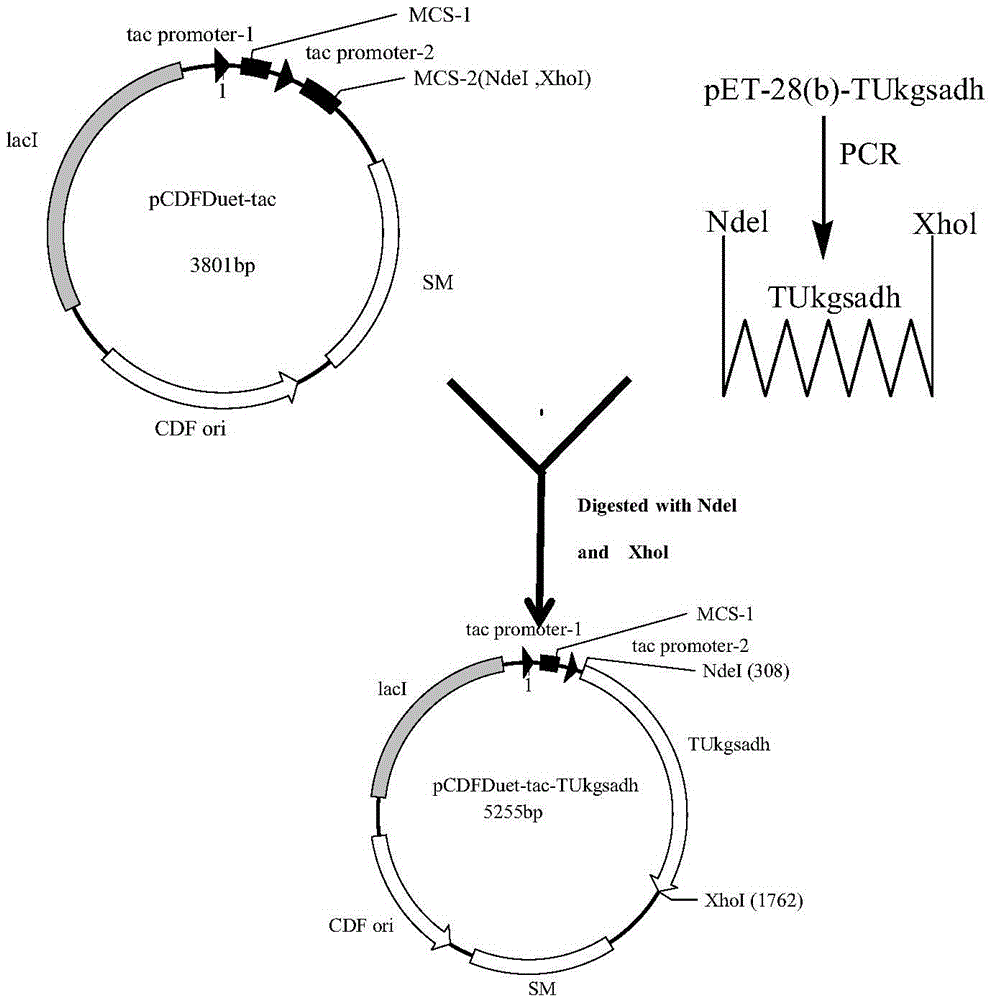
Recombinant Escherichia coli and application of recombinant Escherichia coli in synthesizing 3-hydroxypropionic acid
A technology for recombining Escherichia coli and genes, which is applied in the field of preparation of 3-hydroxypropionic acid, can solve the problems of high price, unbalanced activities of glycerol dehydratase and aldehyde dehydrogenase, complicated construction process, etc., and achieves the effect of increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Replacement of promoters in plasmids
[0050] Strains and plasmids: E.ciliBL21(DE3) was purchased from Transgen, and expression vectors pACYCDuet-1 and pCDFDuet-1 were purchased from Novozymes.
[0051] The replacement of the promoters of plasmids pACYCDuet-1 and pCDFDuet-1 is to replace the sequence of the T7 promoter on the plasmid with the sequence of the tac promoter through homologous recombination.
[0052] Take the construction of pACYCDuet-tac plasmid as an example. Cultivate the Escherichia coli strain containing plasmid pACYCDuet-1, then use the pACYCDuet-1 plasmid as a template, use mutation primers to mutate the T7 sequence on the plasmid template into tac, and then chemically transform the template digested with DpnI endonuclease into BL21( DE3) in competent cells. Then spread on the LB solid medium plate containing 50mg / L chloramphenicol, and PCR screen positive clones. Recombinant plasmids were extracted from positive clones for sequencing v...
Embodiment 2
[0062] 1. Cloning and expression of glycerol dehydratase and glycerol dehydratase activator in Klebsiella pneumoniae
[0063] (1) Bacterial strains and plasmids: Klebsiella pneumoniae (K. peneumoniaeDSM2026) was purchased from DSM Company of Germany, Escherichia coli E.coliBL21 (DE3) was purchased from Transgen Company, and the expression vector pACYCDuet-tac was prepared by the method in Example 1.
[0064] (2) Glycerol dehydratase gene (dhaB123) (dhaB1GeneID: 7947197; dhaB2GeneID: 7947198; dhaB3GeneID: 7947200) and glycerol dehydratase activator gene (gdrA) (gdrAGeneID: 6936977) were cloned using K. peneumoniaeDSM2026 as a template, through routine Amplified by PCR method. (the primer sequence of amplifying glycerol dehydratase gene and glycerol dehydratase activator gene is:
[0065] dhaB1-4-F-EcoR1: CCG GAATTCATGAAAAGATCAAAACGATTTGCAGTACT,
[0066] dhaB1-4-R-HindIII:GTT AAGCTT GATCTCCCACTGACCAAAGCTGG)
[0067] The amplified product was recovered by the Clean-up kit ...
Embodiment 3
[0102] Embodiment 3: Shake flask fermentation experiment of recombinant bacterial strain and recombinant bacterial strain implement the expression of SDS-PAGE target protein
[0103] The recombinant Escherichia coli constructed by the method in Example 2 was inoculated into LB solid medium containing 50 mg / L chloramphenicol and 50 mg / L streptomycin sulfate, and cultured at 37°C for 16 hours to obtain activated recombinant Escherichia coli.
[0104] The activated recombinant Escherichia coli was inoculated into LB medium containing 50 mg / L chloramphenicol and 50 mg / L streptomycin sulfate, and cultured at 37° C. for 10 hours to obtain seed liquid.
[0105] The seed liquid was inoculated into 50 mL of fermentation medium containing 50 mg / L chloramphenicol and 50 mg / L streptomycin sulfate according to the inoculum size of 6% volume concentration, and cultured with shaking at 37° C. and 150 rpm. When OD 600 When it reaches about 0.6, add IPTG to the fermentation broth, the final con...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com