Method of rapidly judging survival state of mycobacterium paratuberculosis in milk and milk product by applying BLU-V PMA technology
A BLU-V, mycobacteria technology, applied in the field of microbial detection, can solve the problems of reduced PMA treatment effect, false positives, short peak brightness time, etc., to achieve the effect of protecting health, rapid development, simple operation, and improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Primer specificity verification
[0031] The genomic DNA of 6 standard strains of Map (P10 and P18), Staphylococcus aureus, Salmonella, Escherichia coli, and Mycobacterium phlei are extracted and amplified by TaqMan fluorescent quantitative PCR to detect the specificity of the method of the invention. (Mycobacterium paratuberculosis ( M.paratuberculosis ) Standard strains: P18 (United States), P10 (Japan) (both are standard strains of bovine paratuberculosis), all provided by Jilin Veterinary Research Institute. Staphylococcus aureus, Salmonella, Escherichia coli, and Mycobacterium phlei are all preserved in the laboratory of the Technical Center of Shanxi Entry-Exit Inspection and Quarantine Bureau. )
[0032] 1. DNA preparation
[0033] Take 50 μL of bacterial solution, add 400 μL of TE, and mix. The specific steps for extracting DNA template are as follows:
[0034] (1) Heat in a water bath at 80 ℃ for 20 min, then cool to room temperature to inactivate;
[0035] (2) Add...
Embodiment 2
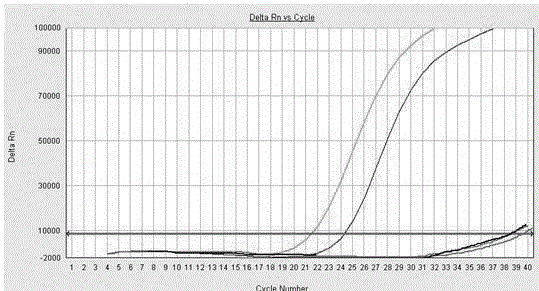
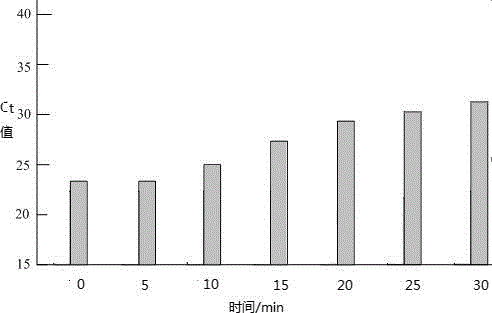
[0048] Optimization of the optimal reaction conditions of PMA on MAP standard strains
[0049] 1. Preparation of MAP standard strains (P10 and P18) membrane damage bacteria
[0050] Take 1 mL of P10 and P18 bacterial solution (take as many bacteria as possible) in a 2 mL EP tube with 50 magnetic beads, seal it with a parafilm, and put it in the nucleic acid extractor (act once every 5 s), Suspend the bacterial liquid thoroughly; after suspension, inactivate it in a water bath at 80 ℃ for 20 min.
[0051] 2. Treatment of MAP standard strains (P10 and P18) by PMA
[0052] Dissolve 0.5 mg of PMA in 1 mL of RNase-free water to prepare a 0.5 mg / mL stock solution of PMA and store at -20°C. Place the inactivated bacterial solution in a centrifuge and centrifuge at 14500 r for 5 minutes; after centrifugation, discard the supernatant; then add 500 μL of EB Buffer to the remaining bacterial cells and mix.
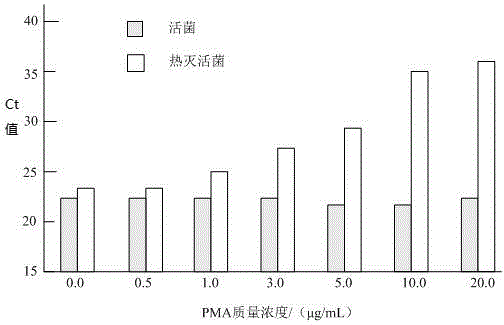
[0053] 3. Optimization of PMA mass concentration
[0054] Add a certain amount of PMA to ...
Embodiment 3
[0060] Quantitative detection of viable bacteria in milk and milk products under optimal PMA reaction conditions
[0061] 1. Preparation of membrane-damaging bacteria (P10 and P18) in artificially contaminated MAP milk samples
[0062] Take 1 mL of P10 and P18 bacterial solution (take as many bacteria as possible) in a 2 mL EP tube with 50 magnetic beads, seal it with a parafilm, and put it in the nucleic acid extractor (act once every 5 s), Suspend the bacterial mass as a bacterial suspension; aseptically take a negative milk sample that has not detected MAP by fluorescent PCR, ordinary PCR and electrophoresis, that is, take 1 mL of yogurt and 0.5 g of milk powder in sterilized 2 mL In the EP tube; add 500 μL of bacterial suspension to the obtained milk sample, mix thoroughly on a vortex shaker; after mixing, place it in a water bath at 80 ℃ for 20 min inactivation. Through this treatment, the cell wall and cell membrane of the bacteria are damaged to different degrees to achieve...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com