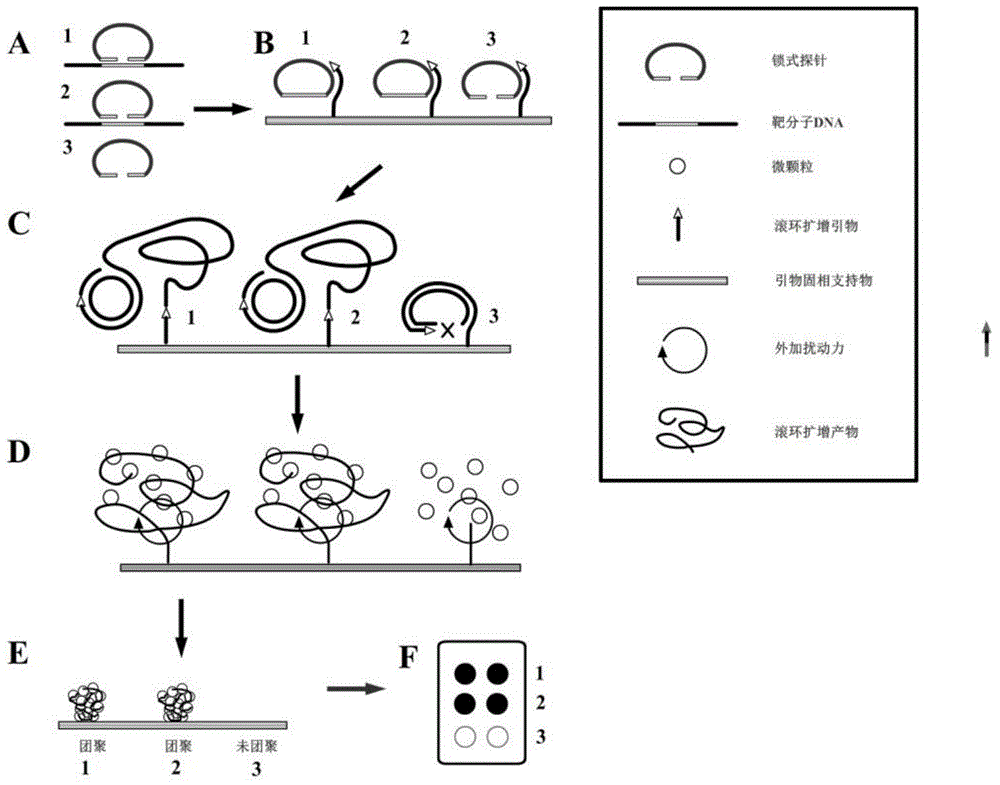
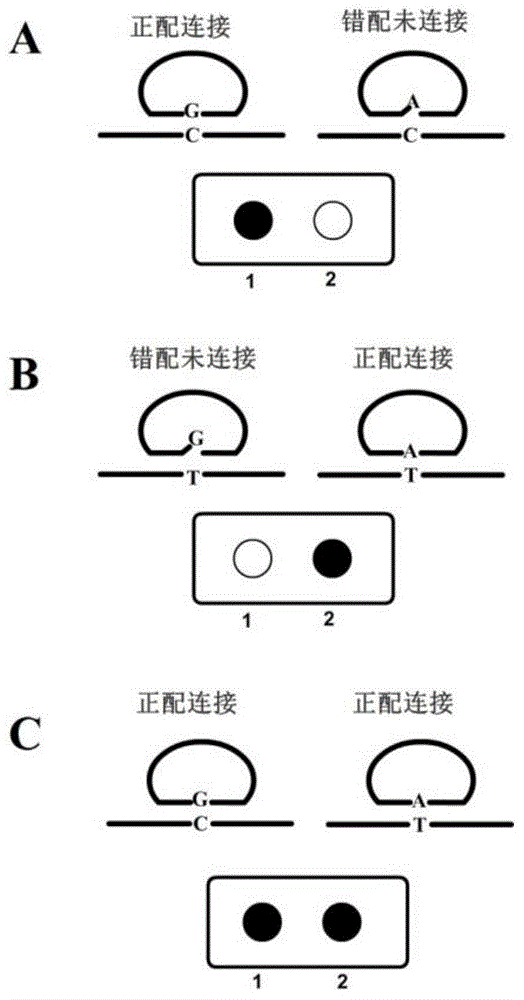
Multiplex nucleic acid visualization detection method and kit based on solid phase rolling circle amplification and particle aggregation
A rolling circle amplification and multiple nucleic acid technology, which is applied in the field of new multiple nucleic acid sequence detection methods and kits, can solve the problems of high equipment and site requirements, and achieve the effects of reducing hardware equipment requirements, fast speed, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
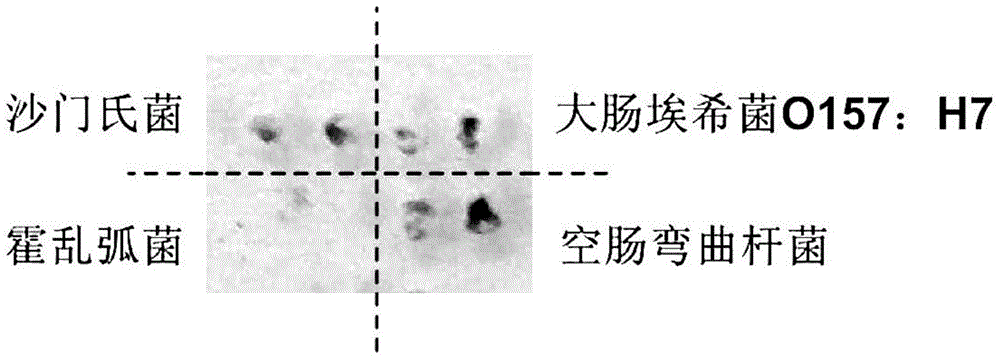
[0070] Example 1: Detection of Foodborne Pathogens
[0071] 1. Samples and reagents
[0072] Genomic DNA of Salmonella, Escherichia coli O157:H7, Vibrio cholerae and Campylobacter jejuni were purchased from ATCC;
[0073] phi29 polymerase and Taq DNA ligase were purchased from NEB (New England Biolabs);
[0074] Other reagents were of domestic analytical grade.
[0075] 2. Primers and probes
[0076] Salmonella invA gene padlock probe sequence such as 5'-SEQ ID NO.1;
[0077] Salmonella amplification primer sequence such as 5'-NH 2 (Cl 2 )-SEQ ID NO.2;
[0078] Escherichia coli O157: H7rfbE gene padlock probe sequence such as 5'-SEQ ID NO.3;
[0079] Escherichia coli amplification primer sequence such as 5'-NH 2 (Cl 2 )-SEQ ID NO.4;
[0080] Vibrio cholerae OMPW gene padlock probe sequence such as 5'-SEQ ID NO.5;
[0081] Vibrio cholerae lock amplification primer sequence such as 5'-NH 2 (Cl 2 )-SEQ ID NO.6;
[0082] Campylobacter jejuni ceuE gene padlock probe s...
Embodiment 2
[0093] Example 2: SNP typing detection of rs 1801133 and rs 1801131 loci in the human genome
[0094] 1. Samples and reagents
[0095] Human genomic DNA was extracted from peripheral blood samples by conventional phenol-chloroform method;
[0096] phi29 polymerase and Taq DNA ligase were purchased from NEB (New England Biolabs);
[0097] Other reagents were of domestic analytical grade.
[0098] 2. Primers and probes
[0099] rs1801133 probe sequence:
[0100] Wild type such as 5'-SEQ ID NO.9;
[0101] Mutant type such as 5'-SEQ ID NO.10;
[0102] Wild-type amplification primer sequence such as 5'-NH 2 (Cl 2 )-SEQ ID NO.11;
[0103] Mutant amplification primer sequences such as 5'-NH 2 (Cl 2 )-SEQ ID NO.12;
[0104] A1298C padlock probe sequence:
[0105] Wild type such as 5'-SEQ ID NO.13;
[0106] Mutant type such as 5'-SEQ ID NO.14;
[0107] Wild-type amplification primer sequence such as 5'-NH 2 (Cl 2 )-SEQ ID NO.15;
[0108] Mutant amplification primer seq...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com