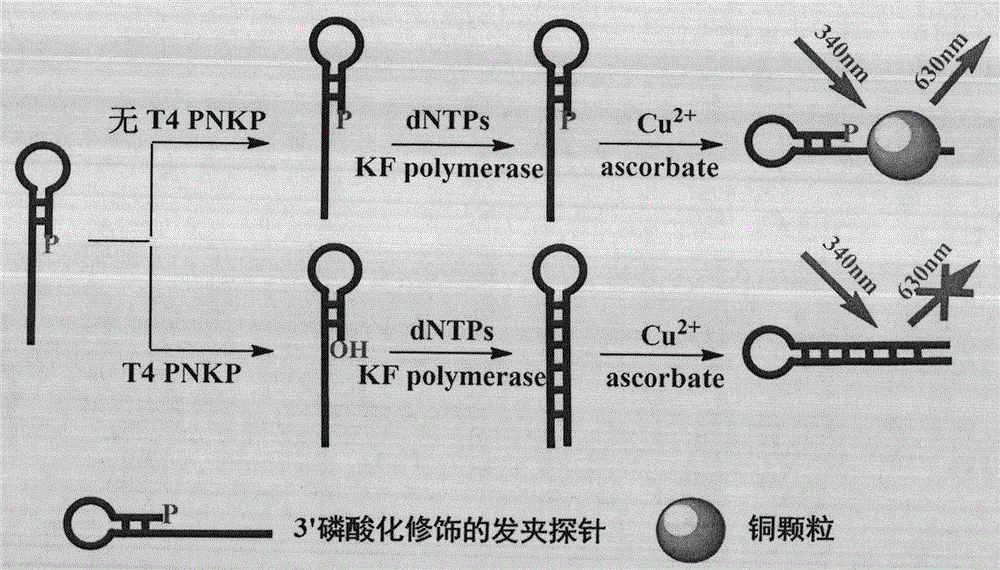
Novel method for beacon-free detection of T4 PNKP (T4 polynucleotide kinase)/phosphatase and inhibitor of T4 PNKP/phosphatase on basis of fluorescent copper nanoparticles
A polynucleotide and nanoparticle technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of application limitation, low selectivity and sensitivity, and high cost, and achieve high sensitivity and selectivity. The effect of good performance and cost reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Step 1. Design the probe sequence as follows: 5′-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT CGC ACC TAA AG G GTG CG -3' (phosphorylation modification at the 3' end);
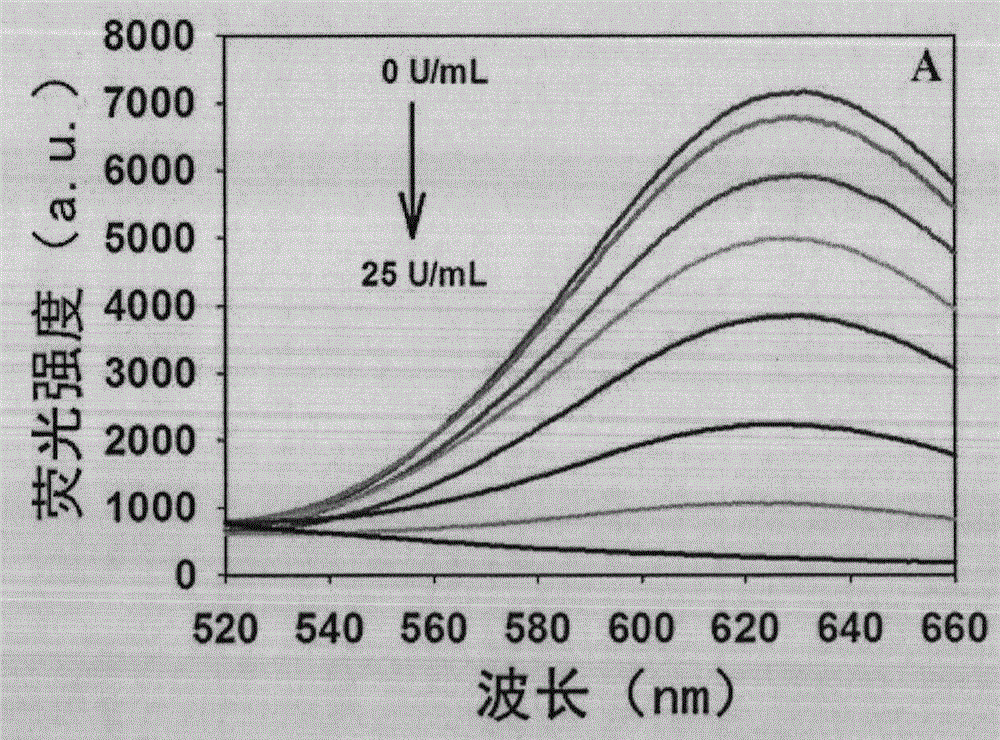
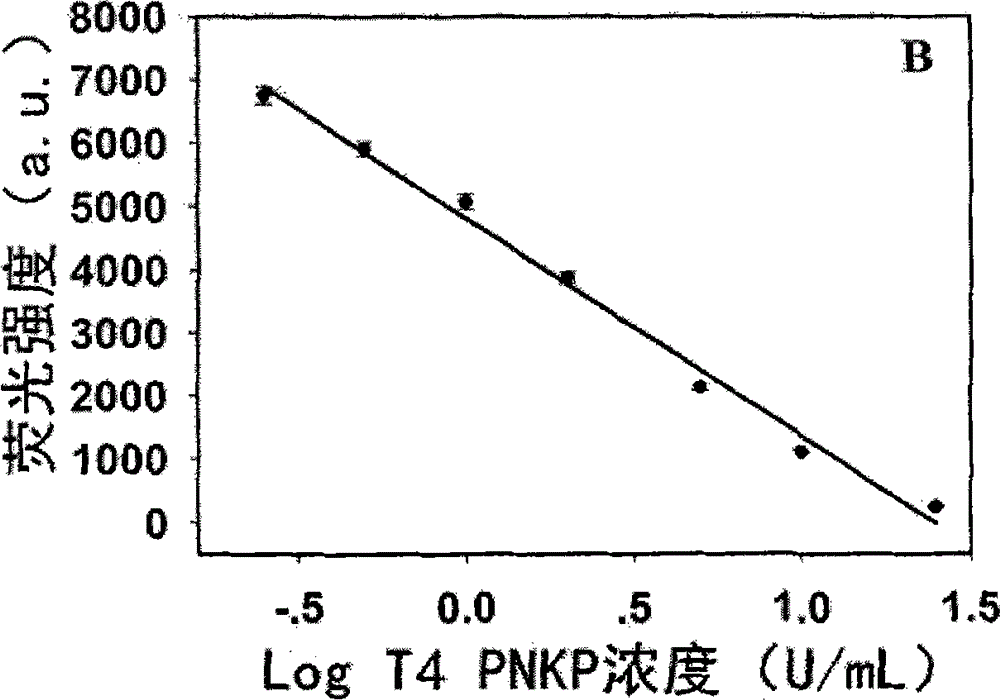
[0041] Step 2. 25 μL Tris-HCl buffer system (10 mM Tris-HCl, 10 mM MgCl, 50 mM NaCl, pH 7.9) contains 5 μL probe (1 μM), 1 μL KF polymerase (50 U / mL), 2 μL dNTPs (200 μM) and 2 μL Different concentrations of T4 PNKP (0U / mL, 0.25U / mL, 0.5U / mL, 1U / mL, 2U / mL, 5U / mL, 10U / mL, 25U / mL) were mixed evenly and kept at a constant temperature of 37°C Incubate for 80min;
[0042] Step 3. Supplement the above solution with a final volume of 100 μL MOPS buffer solution (10 mM MOPS, 150 mM NaCl, pH 7.6), which includes 10 μL sodium ascorbate (2 mM) and 10 μL copper sulfate (200 μM), mix well and incubate at room temperature 10min;
[0043] Step 4, the final reaction solution obtains the fluorescence spectrum figure of 520~660nm under the excitation wavelength of 340nm (see Figure 2a ) and the linear corresponde...
Embodiment 2
[0045]Step 1. Design the probe sequence as follows: 5′-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT CGC ACC TAA AG G GTG CG -3' (phosphorylation modification at the 3' end);
[0046] Step 2. 25 μL Tris-HCl buffer system (10 mM Tris-HCl, 10 mM MgCl, 50 mM NaCl, pH 7.9) contains 5 μL probe (1 μM), 1 μL KF polymerase (50 U / mL), 2 μL dNTPs (200 μM), 5 μL Different concentrations of heparin (0μg / mL, 0.2μg / mL, 0.5μg / mL, 1μg / mL, 2μg / mL, 5μg / mL, 10μg / mL, 25μg / mL) and 2μL T4 PNKP (25U / mL), mixed Evenly, incubate at 37°C for 80 minutes;
[0047] Step 3. Supplement the above solution with a final volume of 100 μL MOPS buffer solution (10 mM MOPS, 150 mM NaCl, pH 7.6), which includes 10 μL sodium ascorbate (2 mM) and 10 μL copper sulfate (200 μM), mix well and incubate at room temperature 10min;
[0048] Step 4, the corresponding relationship between the relative fluorescence intensity of the system and the concentration of heparin is obtained from the final reaction solution at an ...
Embodiment 3
[0050] Step 1. Design the probe sequence as follows: 5′-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT CGC ACC TAA AG G GTG CG -3' (phosphorylation modification at the 3' end);
[0051] Step 2, 25 μL 1% lung cancer cell lysate Tris-HCl buffer system (10mM Tris-HCl, 10mM MgCl, 50mM NaCl, pH 7.9) contains 5 μL probe (1 μM), 1 μL KF polymerase (50 U / mL), 2 μL dNTPs (200μM) and 2μL of different concentrations of T4 PNKP (0U / mL, 0.25U / mL, 0.5U / mL, 1U / mL, 2U / mL, 5U / mL, 10U / mL, 25U / mL), mix well, in Incubate at 37°C for 80 minutes;
[0052] Step 3. Supplement the above solution with a final volume of 100 μL MOPS buffer solution (10 mM MOPS, 150 mM NaCl, pH 7.6), which includes 10 μL sodium ascorbate (2 mM) and 10 μL copper sulfate (200 μM), mix well and incubate at room temperature 10min;
[0053] Step 4, the final reaction solution obtains the fluorescence spectrum figure of 520~660nm under the excitation wavelength of 340nm (see Figure 4a ) and the corresponding relationship ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com