Gene chip and its detection method for identification of six kinds of swine disease pathogens
A technology of gene chips and pathogens, applied in the field of biochips, can solve the problem of lack of parallel detection of six pathogens, and achieve the effect of strong and stable hybridization signal, high sensitivity and strong stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Embodiment 1, the present invention respectively cultures Porcine Infectious Actinobacillus pleuropneumoniae, Haemophilus parasuis and Mycoplasma hyopneumoniae, and extracts pathogen genome DNA; Porcine circovirus type 2, porcine reproductive and respiratory syndrome virus and CSFV cells were propagated, and the virus genome was extracted. Cloning, sequencing and analysis of the specific conserved sequences of Actinobacillus pleuropneumoniae, Haemophilus parasuis, Mycoplasma hyopneumoniae, porcine circovirus type 2, porcine reproductive and respiratory syndrome virus and classical swine fever virus, according to the sequence Results Six oligonucleotide probes were designed, which could specifically bind to the PCR products of corresponding pathogens. When performing genome amplification by PCR, the 5′ end of the upstream universal primer was labeled with Cy3 fluorescent dye, and the probes could bind to the PCR products. Hybridization was performed at 42°C. According t...
Embodiment 2
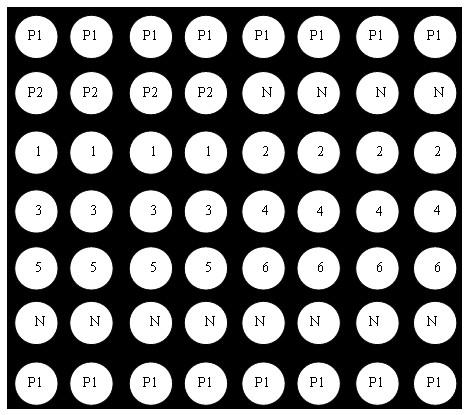
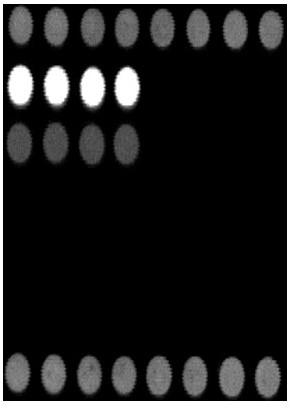
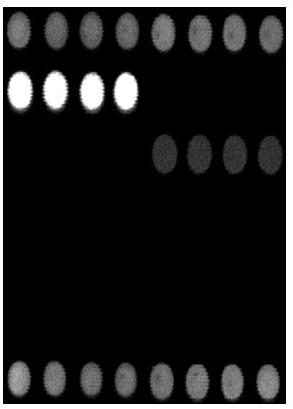
[0081] Example 2, see figure 1 , the gene chip used for the detection of important pathogens of respiratory disease syndrome, adopts the micro-spotting technology of the gene chip, immobilizes the probes of the samples to be tested and various quality control probes on the chemically modified substrate, forming 8 rows × 7 columns microarray. The distribution of probes on the microarray from top to bottom is as follows: spotting position quality control partition P1: 1 row × 8 points, hybridization positive quality control partition P2: 1 row × 4 points, hybridization negative quality control partition N: 1 row × 4 o'clock, porcine infectious Actinobacillus pleuropneumoniae detection probe division 1: 1 row × 4 dots, Haemophilus parasuis detection probe division 2: 1 row × 4 dots, Mycoplasma hyopneumoniae detection probe division 3: 1 Row × 4 dots, porcine circovirus type 2 detection probe division 4: 1 row × 4 dots, porcine reproductive and respiratory syndrome virus detectio...
Embodiment 3
[0084] Embodiment 3, reagent preparation
[0085] 1) Chip washing solution
[0086] According to needs and actual conditions, prepare washing solution I and washing solution II according to the following proportions.
[0087] Washing solution Ⅰ: the final concentration of SSC is 2×, and the final concentration of SDS is 0.2%. For example, 600 mL of washing liquid Ⅰ=528 mL of distilled water+60mL of 20×SSC+12mL of 10%SDS, or prepare in proportion as required.
[0088] Washing solution Ⅱ: The final concentration of SSC is 0.2×. For example, 600 mL of washing solution II=594 mL of distilled water+6 mL of 20×SSC, or prepare in proportion as required.
[0089] If 10% SDS produces a white flocculent precipitate, please dissolve it in a water bath at 42°C and mix well to prepare a lotion.
[0090] 2) Absolute ethanol.
[0091] 3) Ice-water mixture.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com