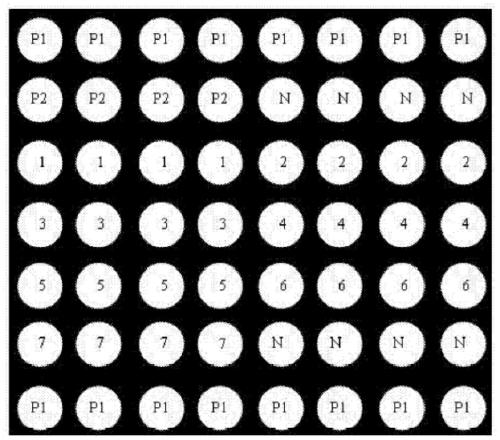
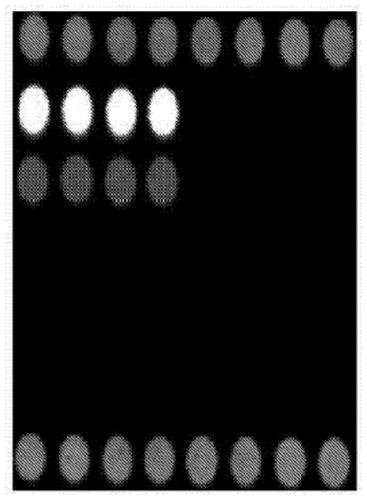
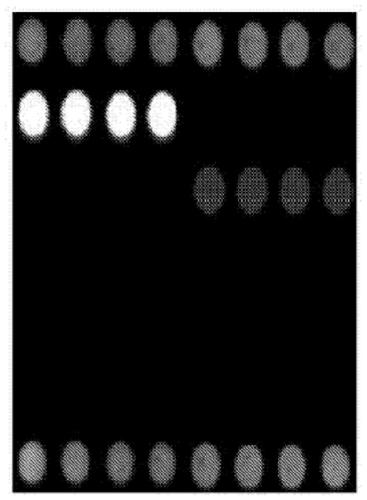
Gene chip for identifying seven fungal infection pathogens as well as kit and detection method thereof
A gene chip and fungal infection technology, applied in the field of gene chips, can solve the problems of time-consuming and labor-intensive, false positives, existence of pollution, etc., and achieve the effects of stability, time-saving and labor-saving, stable hybridization signal, and strong hybridization signal.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Extraction of sample DNA to be tested
[0048] Extraction can be carried out according to conventional methods in the field, and samples can be fresh tissue fluid, blood, urine, cerebrospinal fluid, secretions (sputum, pleural effusion, ascites, alveolar lavage fluid, etc.). In this embodiment, Qiagen micro DNA extraction kit is used, taking fresh tissue as an example:
[0049] (1) Put the fresh tissue into a sterile mortar, pour it into liquid nitrogen to freeze and grind it, then transfer the ground tissue into a 1.5mL EP tube;
[0050] (2) Add 1 mL of LPBS buffer to wash, centrifuge at 3000 rpm for 5 minutes, and discard the supernatant;
[0051] (3) Add 200 μL of 50 mM NaOH solution, incubate at 95°C for 10 minutes, centrifuge at 5000 rpm for 10 minutes, and discard the supernatant;
[0052] (4) Add 500 μL Lyticase solution, incubate at 37°C for 30 minutes, centrifuge at 14,000 rpm for 10 minutes, and discard the supernatant;
[0053] (5) Quickly add 18...
Embodiment 2
[0062] Example 2 PCR amplification of the fungal rDNA-ITS sequence of the sample to be tested
[0063] The PCR reaction system includes: rTaq enzyme 12.5 μL, primer Mix 2 μL, test sample DNA / cDNA 5 μL, nuclease-free sterilized water 5.5 μL;
[0064] Wherein, the primer Mix contains: the sequence is SEQ ID NO: 10 20 μmol / L fungal gene amplification (Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, Candida krusei, Aspergillus fumigatus and Aspergillus flavus) universal primer upstream primer 1 μ L, the sequence is 20 μ mol / L fungal gene amplification of SEQ ID NO: 11 (Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, Candida krusei, Aspergillus fumigatus Mold and Aspergillus flavus) universal primer downstream primer 1 μL.
[0065] SEQ ID NO: 10:
[0066] 5'-TCCGTAGGTGAACCTGCGG-3';
[0067] SEQ ID NO: 11:
[0068] 5'-TCCTCCGCTTATTGATATGC-3';
[0069] Amplify according to the following steps: 94°C 5min; 94°C 30sec, 58°...
Embodiment 3
[0070] Example 3 Design of Fungal Specific Probes
[0071] The present invention carries out sequence analysis to the genome DNA of Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, Candida krusei, Aspergillus fumigatus and Aspergillus flavus, and designs 7 oligonucleotide probes. The needle can specifically bind to the PCR product (rDNA-ITS) of the corresponding pathogen. When performing genome amplification by PCR, the 5' end of the upstream universal primer is labeled with Cy3 fluorescent dye, and the probe can be carried out with this product at 42°C. hybridize. According to the position and intensity of the fluorescent signal, the hybridization result is judged to identify Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, Candida krusei, Aspergillus fumigatus and Aspergillus flavus infection. According to the complete gene sequences of Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, Can...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com