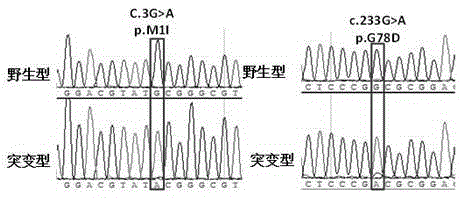
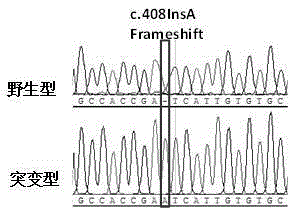
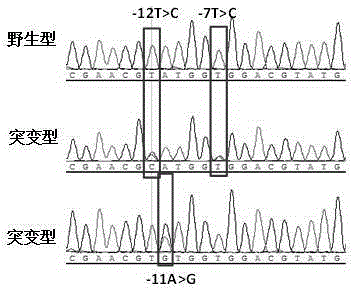
Kit for detecting mycobacterium tuberculosis pncA gene mutation
A Mycobacterium tuberculosis and kit technology, applied in the direction of microorganism-based methods, microorganism measurement/testing, microorganisms, etc., can solve the problems of public reports of undetected mutation sites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1: collecting detection samples
[0048] 16 strains of pyrazinamide-resistant tuberculosis strains were collected, and the clinical patients were all pulmonary tuberculosis patients. According to the test results of drug-susceptible pyrazinamide (PZA content 1 μg / ml), a total of 16 strains of pyrazinamide-resistant tuberculosis strains were obtained. The mean age of patients resistant to pyrazinamide was 40 years. Male patients accounted for 68.75% (11 / 16), aged between 15-69 years old, with an average age of 40.3 years; female patients accounted for 31.25% (5 / 16), aged between 21-49 years old, with an average age of 39.4 age.
Embodiment 2
[0049] Example 2: Extraction of Genomic DNA
[0050] 1. Use a disposable inoculation loop to scrape the cultured colony of Mycobacterium tuberculosis and place it in a 1.5ml EP tube (try not to scrape the culture medium);
[0051] 2. Add 500 μl of cell suspension to the centrifuge tube of the bacterial sediment (first check whether Lysozyme has been added), use a pipette or a vortex shaker to thoroughly suspend the tuberculosis cell pellet, and incubate at 37°C for 30 minutes and invert every 5-10 minutes Mix several times. Centrifuge at 12000rpm (~13400×g) for 2min, and try to absorb the supernatant;
[0052] 3. Add 225 μl buffer A to the cell pellet, shake until the cell is completely suspended;
[0053] 4. Add 10 μl proteinase K solution to the tube and mix by inverting;
[0054] 5. Add 25 μl lysis buffer S, invert and mix; place in a water bath at 57°C for 20 minutes, and invert and mix several times during this time.
[0055] 6. Add 250μl buffer B, shake for 5s and ...
Embodiment 3
[0062] Embodiment 3: PCR amplification, electrophoresis result
[0063] Using the extracted whole genome DNA of Mycobacterium tuberculosis as a template, PCR amplification was carried out, and the amplified gene was pncA gene, and the primers used were 5'-TGTCGCTCACTACATCACC-3' and 5'-TCGTAGGTCATAGCGTAGG-3'.
[0064] PCR amplification system: 25 μL of 2×PCR master mix (containing rTaq enzyme, TAKARA), 1 μM of forward and reverse primers, 50 ng of template DNA, and 21 μL of deionized water.
[0065] The PCR reaction conditions are: denaturation at 94°C for 5 minutes, followed by 35 cycles (denaturation at 94°C for 30 seconds, annealing at 50°C for 30 seconds, extension at 72°C for 1 minute and 30 seconds), and finally extension at 72°C for 5 minutes. The length of the amplified product is 867bp, and then it is detected by agarose gel electrophoresis. The DL2000 model DNAmarker is used as the control of the PCR amplified product, and the agarose gel with a gel concentration of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com