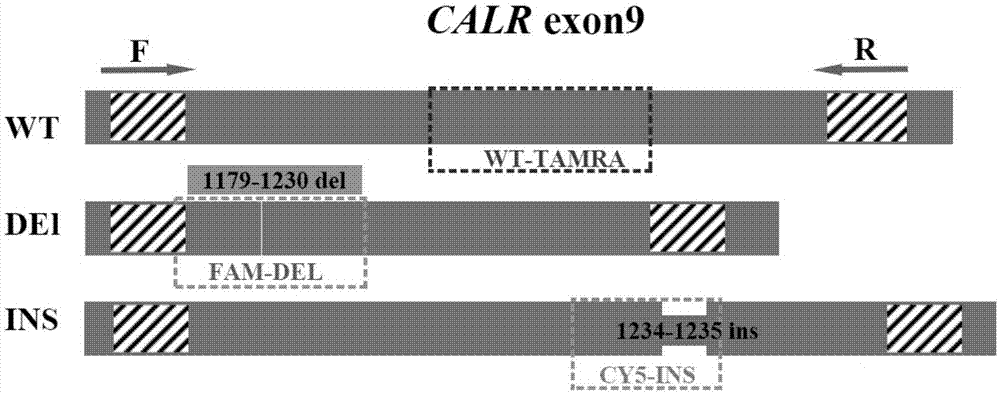
Detection method for CALR (Calreticulin) gene deletion and insertion mutation and kit
An insertion mutation and gene deletion technology, applied in the field of biomedical testing, can solve problems such as CALR gene deletion and lack of diagnostic molecular markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0118] Verify the method of the present invention with positive plasmid sample
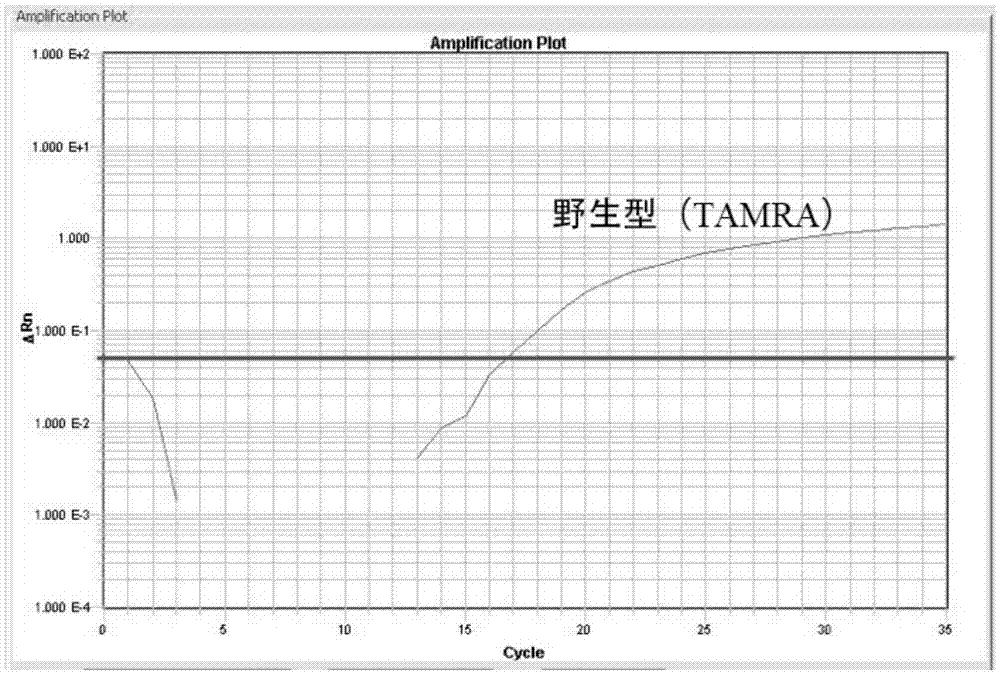
[0119] In this example, the method of the present invention was used to perform PCR detection on each positive known CALR genotype plasmid, and it was verified that the reaction system of this method can identify the wild type, insertion mutation and / or deletion mutation of CALR at one time through multiple reactions in a single tube. Each reaction contained the wild-type, deletion or insertion mutation-positive plasmids of the exon region of CALR9, and the positive plasmids of each CALR genotype were prepared individually or in combination as samples to be tested (plasmid concentration was 10 3 copies / μL).
Embodiment approach
[0121] 1. Prepare standard positive plasmids for each genotype of CALR
[0122] 1.1. Primer design: Design primers according to the reported exon 9 sequence of the human CALR gene:
[0123] Upstream primer 5'-3' sequence: CTGGTCCTGGTCCTGATGTCG
[0124] Downstream primer 5'-3' sequence: TCTTCCTCCTTGTCCTCCTCA
[0125] 1.2. Use the following PCR reaction system to clone the CALR exon 9 mutation region:
[0126] Add the following PCR system as shown in Table-2 into the 50 μL micro-PCR reaction tube:
[0127] Table 2
[0128]
[0129]
[0130] Add sterilized deionized water to a final volume of 50 μl.
[0131] The reaction conditions were total denaturation at 94°C for 5 minutes, 40 cycles (94°C for 30 seconds, 54°C for 1 minute, 72°C for 1 minute), and 72°C for 10 minutes.
[0132] 1.3.PCR product cloning: the amplified PCR fragment was recovered by the PCR product recovery kit, blunt-ended with T4DNA polymerase (T4DNApolymerase, NEBBioLabs company), and then subjected ...
Embodiment 2
[0149] Confirmatory testing of the method of the invention with known CALR genotype blood samples
[0150] The present invention verifies and measures known CALR genotype blood samples
[0151] In this example, for the serum samples with known CALR genotypes after sequencing, PCR verification was performed using the kit of the present invention. This example further proves that this kit is suitable for the identification and detection of the insertion and deletion mutations of exon 9 of the CALR gene in the human serum genome, and is suitable for the diagnosis of MPN related to human CALR.
[0152] 1. Implementation method:
[0153] 1. After labeling the blood samples (27 cases in total) with known CALR gene sequences to be tested, take 150-500 μL of blood, extract DNA with QIAGEN or other corresponding company blood DNA extraction kits, and measure the 260nm O.D. value , and dilute it to a concentration of approximately 5-10 ng / μL.
[0154] 2. The real-time PCR reaction sy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com