A high-expression promoter derived from Kluyveromyces marx and its expression system
A high-expression, promoter technology, applied in microorganism-based methods, using vectors to introduce foreign genetic material, peptides, etc., can solve the problem of limited expression vectors, achieve high transcription activity, fast and convenient screening, and strong transcription initiation activity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] (1) Acquisition of KmINU1 promoter
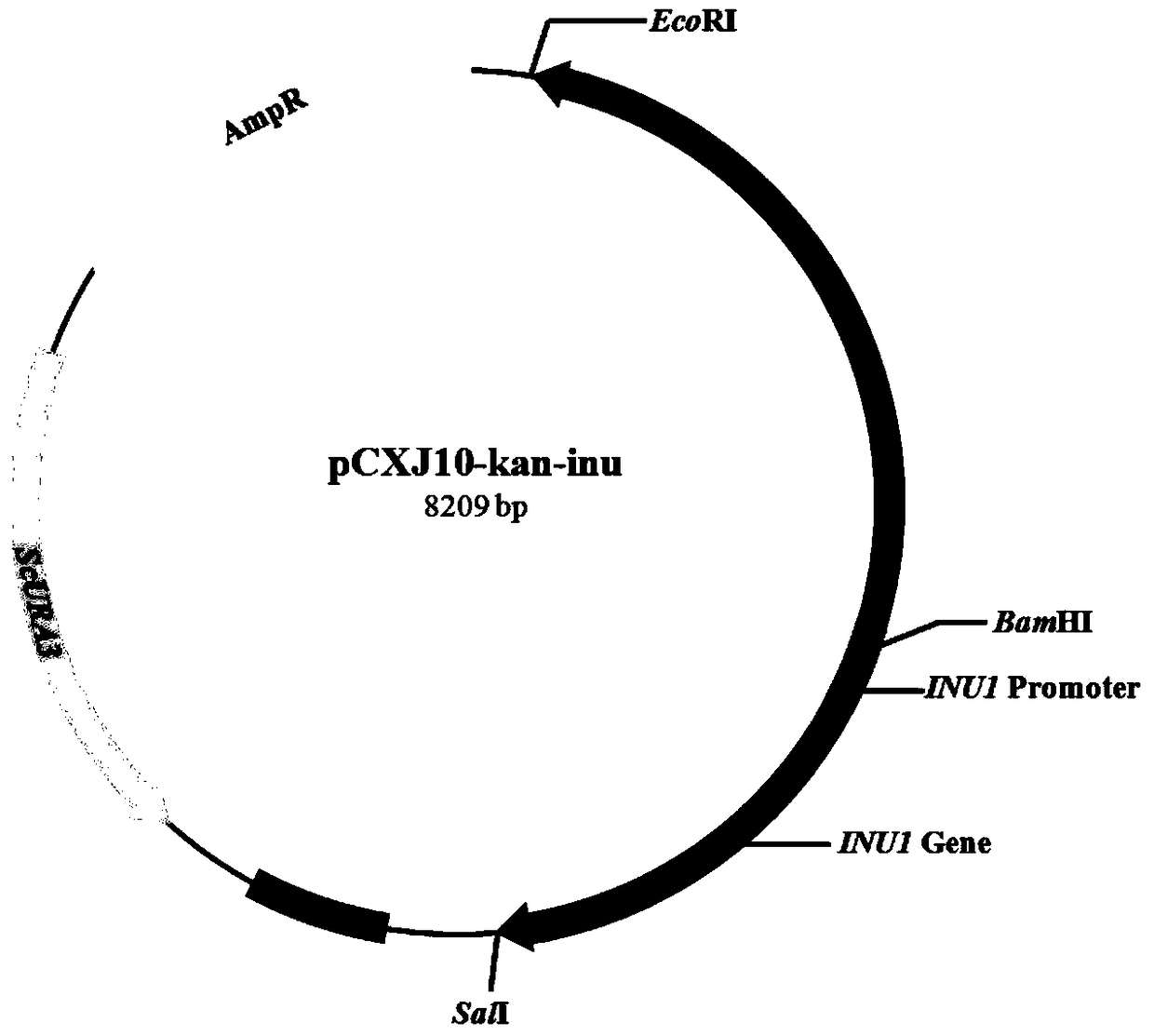
[0046] Using the genome of Kluyveromyces cicerisporus CBS4857 as a template, Inu-1058 (SEQ ID No.11) and Inu-0 (SEQ ID No.13) as primers, the polymerase chain reaction ( PCR) amplification method to obtain the KmINU1 promoter, its nucleotide sequence is shown in SEQ ID No.1, and its total length is 1058bp.
[0047] The genome of Kluyveromyces cicerisporus (Kluyveromyces cicerisporus) CBS4857 was used as a template, and inu-PS-F (SEQ ID No.5) and Inu-0 (SEQ ID No.13) were used as primers. The KmINU1 promoter was obtained by reaction (PCR) amplification, its nucleotide sequence is shown in SEQ ID No. 2, and its total length is 367 bp.
[0048] (2) Acquisition of efficient exocrine signal peptide (SEQ ID No.3)
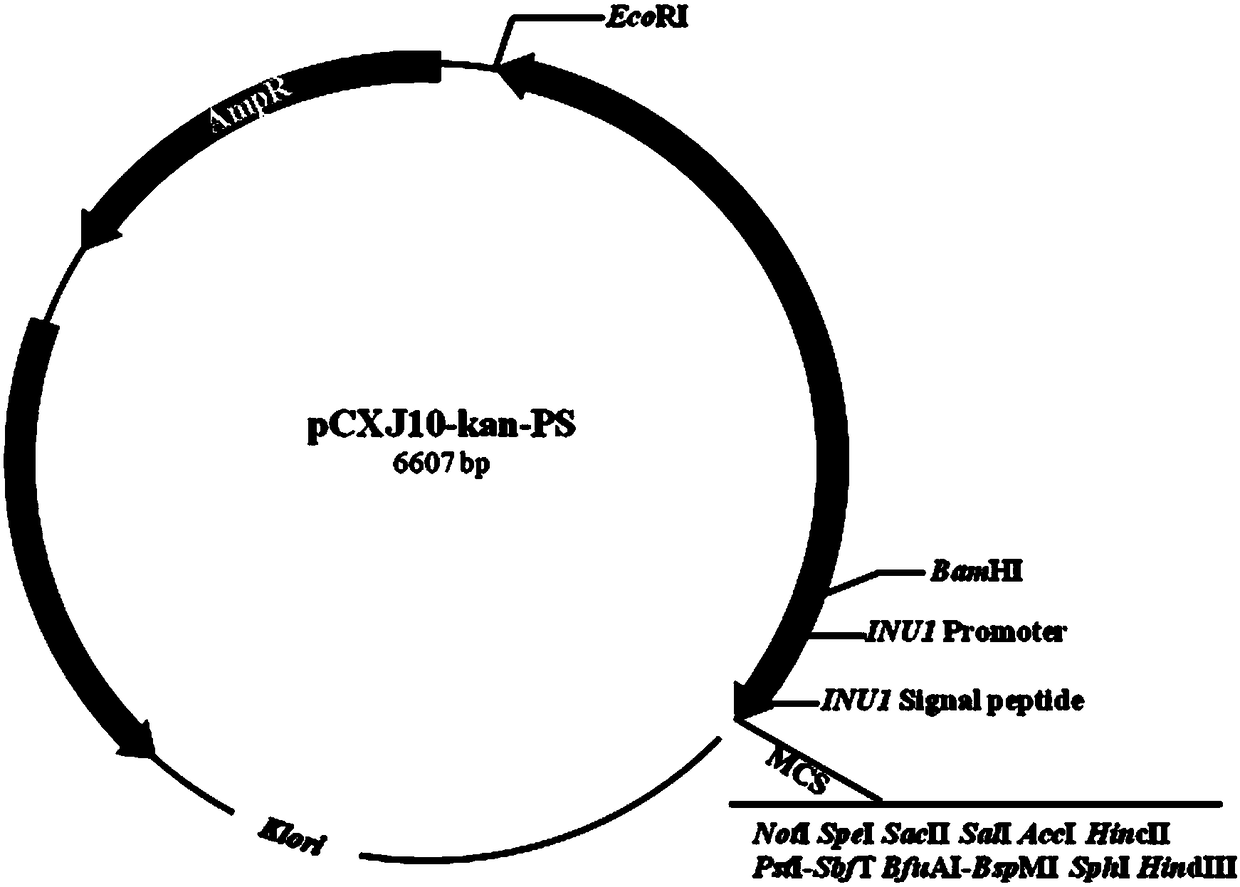
[0049] Using the genome of Kluyveromyces cicerisporus CBS4857 as a template, inu-PS-F (SEQ ID No.5) and inu-PS-R (SEQ ID No.6) as primers, polymerase The inulinase promoter was obtained by chain reaction (PCR) amplification, ...
Embodiment 2
[0052] 1. Differential analysis of the activation activity of KmINU1 promoters with different lengths
[0053] The KmINU1 promoter of SEQ ID No.1 that embodiment 1 obtains is truncated into 5 sections, as Image 6 shown. Image 6 In the above, the transcription start site (ATG) is defined as +1 position, the interval from -1058 to -1 is the KmINU1 promoter, and the interval from +1 to +69 is the high-efficiency exocrine signal peptide of the present invention. The KmlINU1 promoter starts from ATG and is truncated at -217bp, -367bp, -567bp, -817bp, -1058bp, respectively, to obtain KmlINU1 promoters with nucleotide sequence lengths of 217, 367, 567, 817 and 1058bp respectively sequence.
[0054] In order to study the differences in the promoter activities of the above-mentioned KmINU1 promoters of different lengths, and further determine their core promoter regions, the above-mentioned promoters of different lengths were linked to kanamycin resistance gene and reporter gene gr...
Embodiment 3
[0062] 367bp length KmINU1 promoter (shown in SEQ ID No.2) and KmPGK promoter activity comparison
[0063] In order to compare the most ideal KmINU1 promotor (shown in SEQ ID No.2) proposed by the present invention and the strong and weak relationship of the initiating activity of the Kluyveromyces strongest KmPGK promotor reported at present, take Kluyveromyces lactis as Expression host, green fluorescent protein as the reporter gene for experiments. The specific experimental steps are as follows:
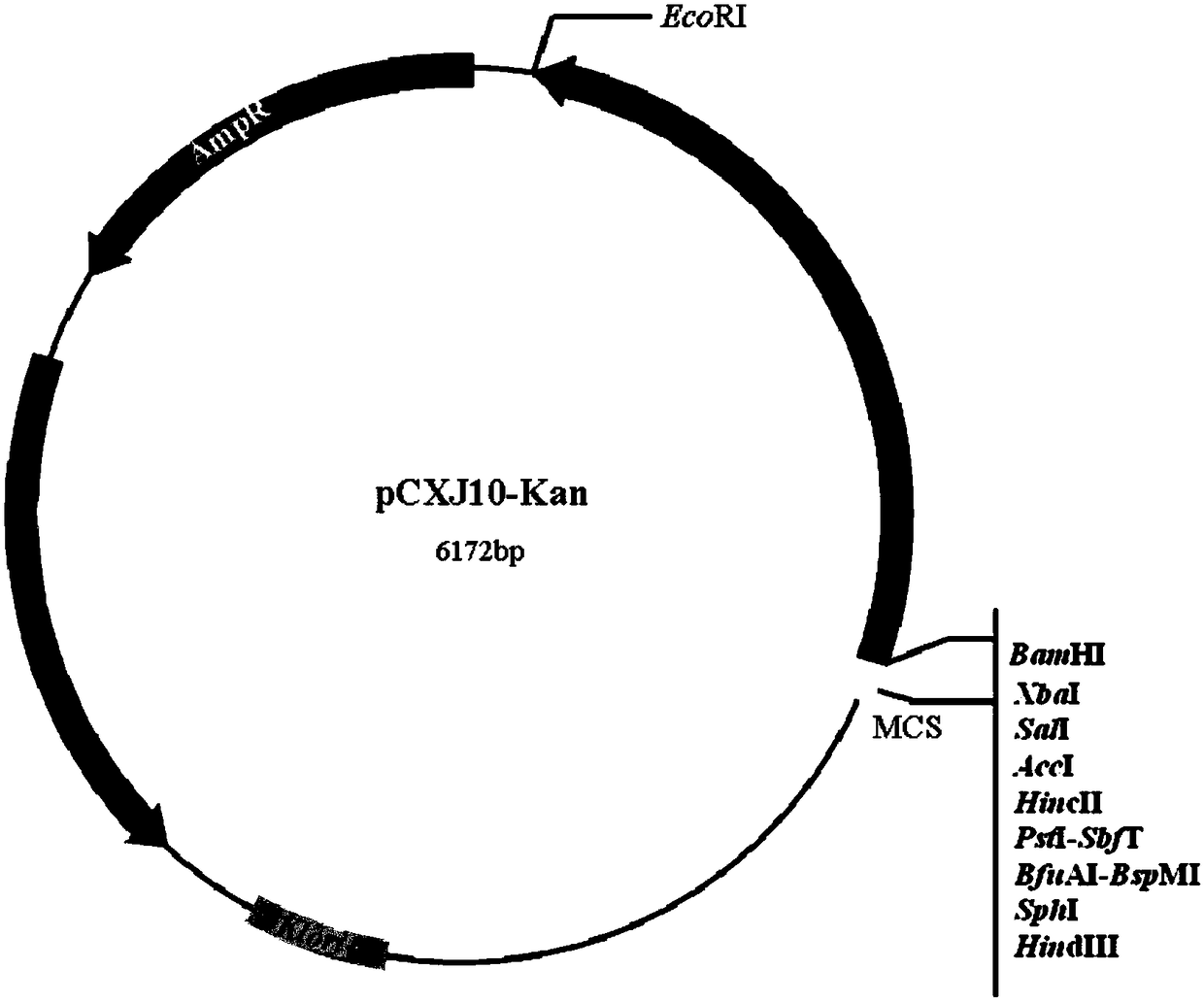
[0064] (1) Using the plasmid pCXJ10 as the basic backbone (GenBank U34314), and according to the characteristics of the restriction site, select BamHI and EcoRI restriction sites to insert the kanamycin resistance gene (kanMX4). The kanMX4 fragment is derived from the double digestion product of plasmid pFA6a-kanMX4, the restriction site is BamHI and EcoRI, the digestion product is recovered and purified to obtain the kanMX4 DNA fragment, which is ligated with the vector pCXJ10 b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com