Primer combination for detecting human EGFR, KRAS and BRAF gene mutation and kit thereof
A kit and human detection technology, which is applied in DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problem that the site involved in the mutation is not the same site, cannot detect new mutations, and is not the same gene and other issues, to achieve the effect of optimizing primer design, reducing non-specific interference, and ensuring consistency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1: Design and Screening of ARMS Primers for Detecting Human EGFR, KRAS, and BRAF Gene Mutations
[0062] When designing ARMS primers in the present invention, Taq enzyme lacks 3'-5' exonuclease activity. When the 3' end of the primer cannot be completely matched with the template, PCR amplification cannot be performed. The 3' end of the primer is designed to only match the mutant template, but not the normal template. The primer can only amplify the mutant template, but not the normal template, so as to achieve the purpose of typing.
[0063] The present invention also designs primer sequences for amplifying normal gene sites corresponding to mutation sites of human EGFR, KRAS, and BRAF genes, and uses the amplified normal gene sites as the second internal reference.
[0064] In this example, ARMS primers for human EGFR, KRAS, and BRAF gene mutations were designed through optimized screening. The sequences are as follows:
[0065] EGFRExon-18-F-WT-ARMS: 5'-CTG...
Embodiment 2
[0100] Example 2: Specific investigation of ARMS primers for detecting human EGFR, KRAS, BRAF gene mutations
[0101] The positive samples containing EGFR, KRAS, and BRAF gene mutations were respectively used as detection objects, and the ARMS primers screened in Example 1 were used for detection.
[0102] The detection reaction system is: TaqHSDNA polymerase (1unit);
[0103] TaqHS DNA polymerase 10× buffer;
[0104] SYBR-Green-I (1:30000);
[0105] ARMS primers (0.1–0.5uM);
[0106] Template DNA (1-10ng);
[0107] 4 kinds of dNTP mixture (0.2mM);
[0108] High purity water.
[0109] PCR reaction conditions:
[0110] Select the SYBR fluorescence channel mode, and the fluorescent quantitative PCR reaction procedure is as follows:
[0111]
[0112] Note: *, collect fluorescence at this step.
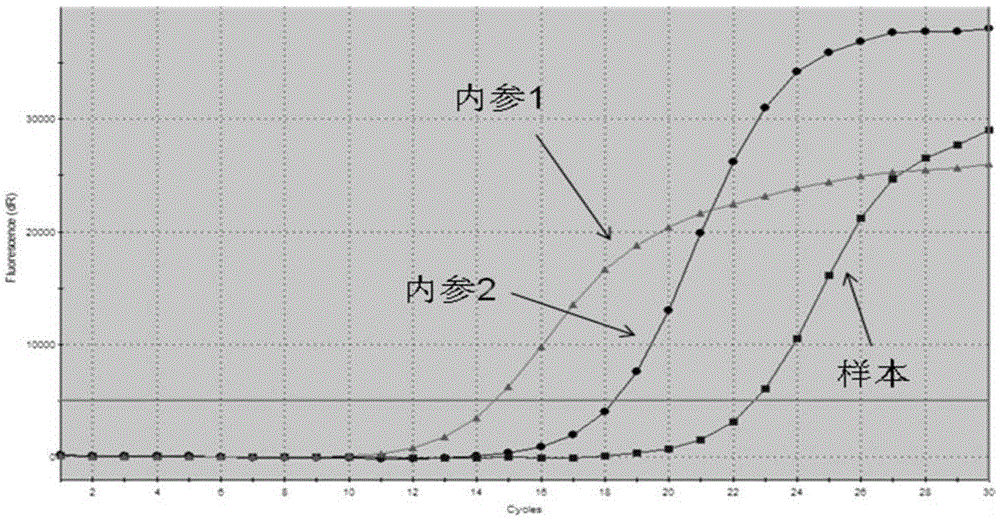
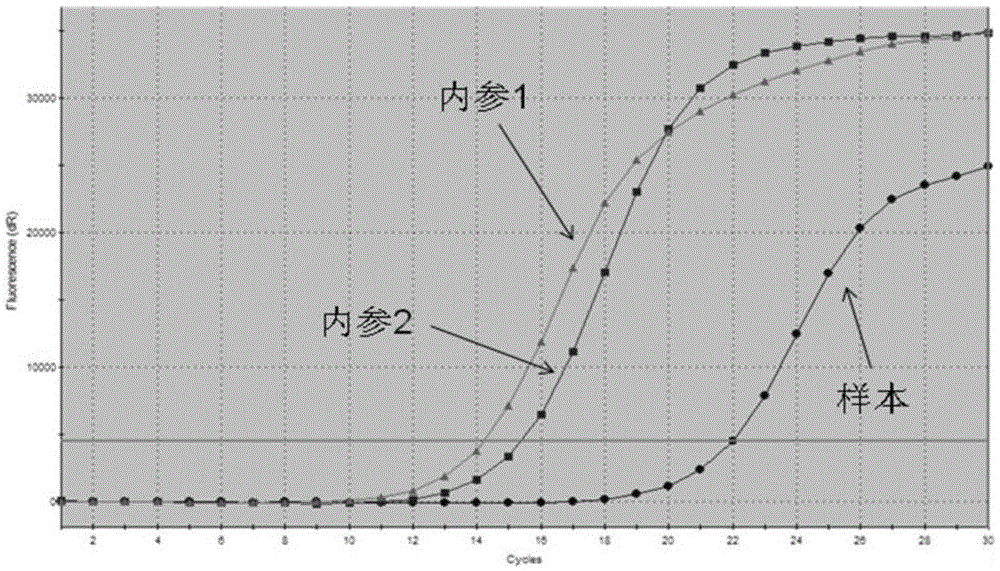
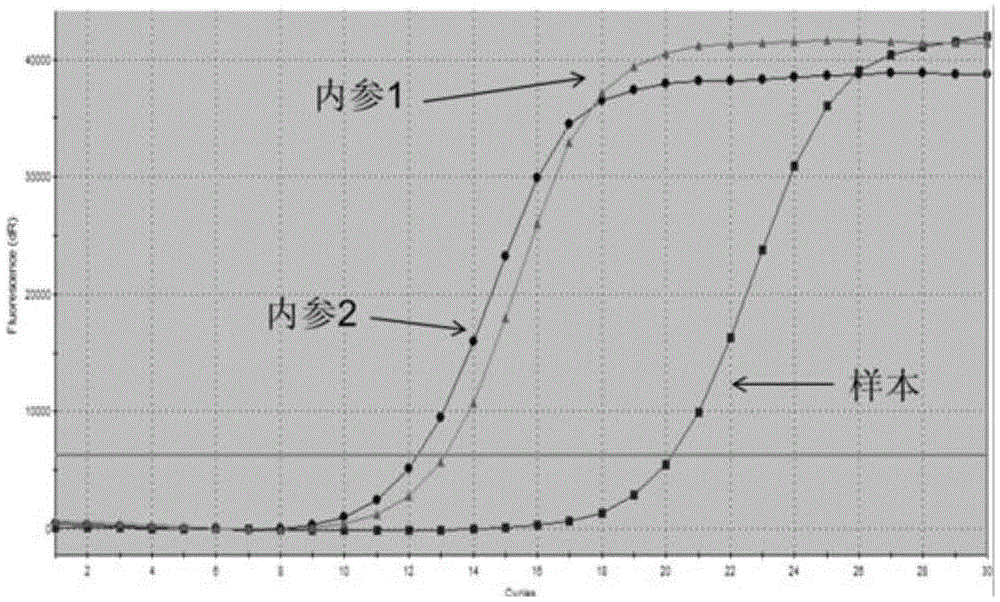
[0113] The results were: the ARMS primers used for detecting EGFR gene mutations could only specifically amplify positive samples containing EGFR gene mutations, but had no amp...
Embodiment 3
[0123] Example 3: Sanger sequencing primers for amplifying EGFR, KRAS, BRAF genes
[0124] The invention also provides Sanger sequencing primers for amplifying EGFR, KRAS, and BRAF genes, including amplification primers and sequencing primers.
[0125] Wherein, the pair of amplification primers used to amplify the region of exon 18 of the EGFR gene is:
[0126] EGFRExon-18-F:5'-CCGCTCGAGCTGAGGTGACCCTTGTCTC-3' (SEQ ID NO.34);
[0127] EGFRExon-18-R:5'-CGGGATCCCCAGACCATGAGAGGCCCTG-3' (SEQ ID NO.35);
[0128] The pair of amplification primers used to amplify the region of exon 19 of the EGFR gene is:
[0129] EGFRExon-19-F:5'-GCCAGTTAACGTCTTTCCTTCTC-3' (SEQ ID NO.36);
[0130] EGFRExon-19-R:5'-GATGTGGAGATGAGCAGGGTC-3' (SEQ ID NO.37);
[0131] The pair of amplification primers used to amplify the region of exon 20 of the EGFR gene is:
[0132] EGFRExon-20-F:5'-AAGCCACACTGACGTGCC-3' (SEQ ID NO.38);
[0133] EGFRExon-20-R:5'-CCTGATTACCTTTGCGATCTGC-3' (SEQ ID NO.39);
[0134] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com