Detection method by using PRSS2 gene as lactoprotein content molecular marker
A molecular marker, detection method technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effect of improving accuracy and precision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] 1. Test materials and methods:
[0035] 1 Experimental materials:
[0036] 1.1 Test animals:
[0037] Taking the Chinese Holstein dairy cow population as the test object, the cows in a large dairy farm in Liaoning Province were selected, and the samples were collected by jugular vein blood sampling.
[0038] 1.2 Test reagents:
[0039]Agarose (BIOWEST); TAE and diethyl pyrophosphate (DEPC) were purchased from Sigma Company; PCR primers were synthesized by Shanghai Bioengineering Company; DNA extraction kit and 2XTaqPCR MasterMix were purchased from TIANGEN Company.
[0040] 1.3 DNA analysis software:
[0041] PCR primer design software: PrimerPremier5;
[0042] Data analysis software: SPSS13.0;
[0043] 2. Experimental method:
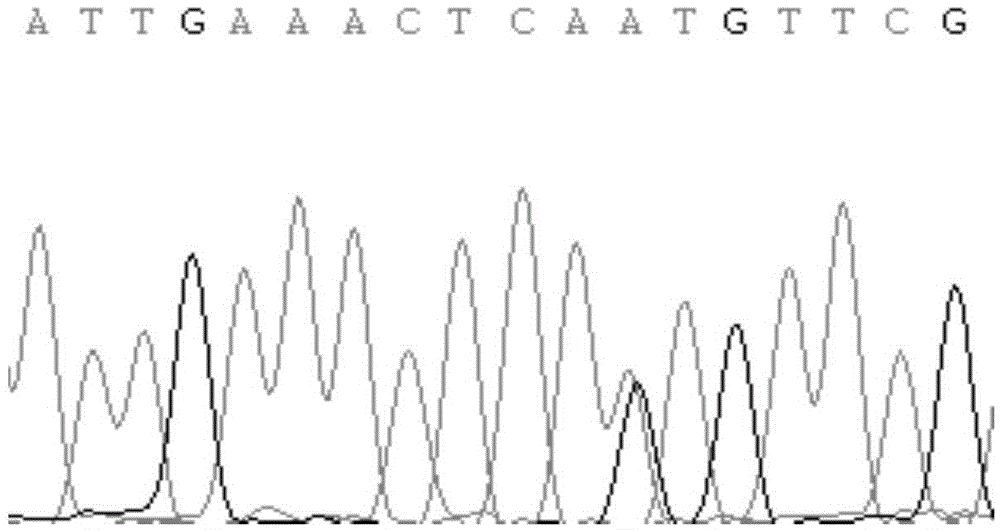
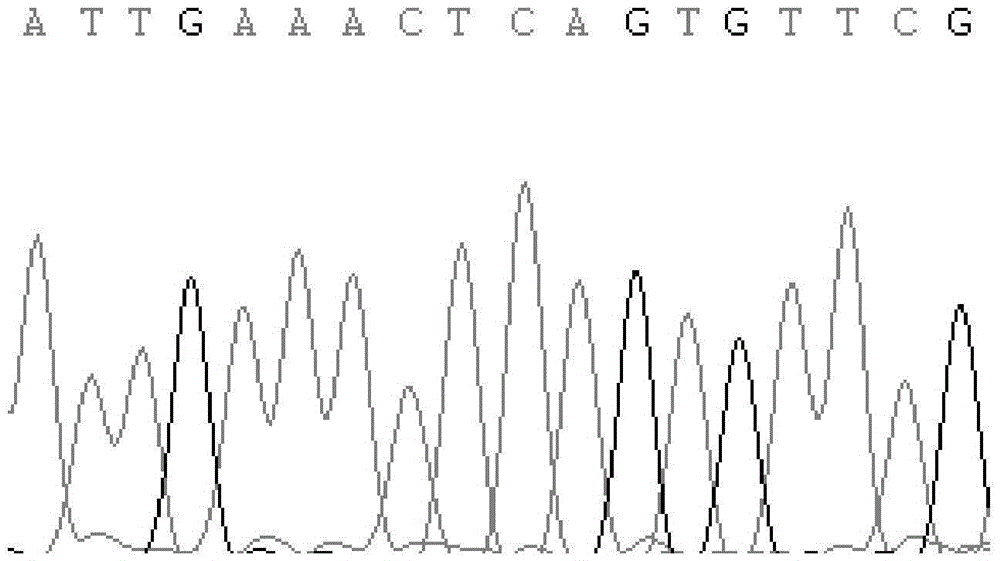
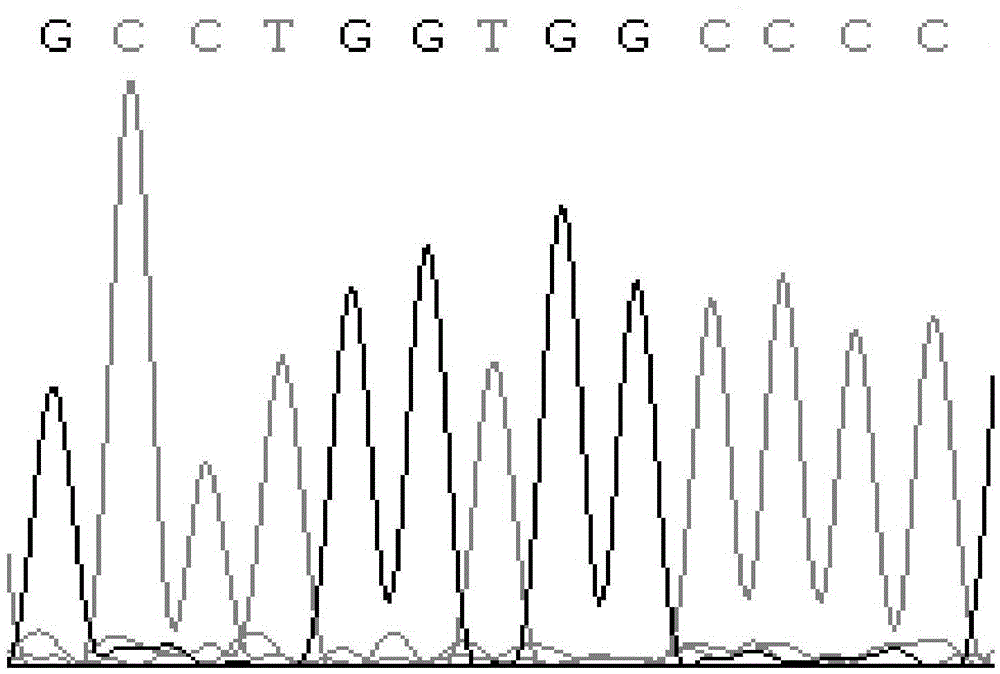
[0044] In this experiment, DNA extraction kit from TIANGEN Company was used to extract the whole genome DNA from the blood of Chinese Holstein cows. At the same time, primers were designed for PCR amplification, and the products were seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com