Anti-PCV (porcine circovirus) diseased pig screening SERPINA1 molecular marker breeding method and application thereof
A porcine circovirus disease and assisted breeding technology, applied in the field of molecular genetics, can solve problems such as elevated serum levels, aggravated lung tissue inflammation, and decreased serum levels
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Cloning of porcine SERPINA1 coding region sequence and search for mutation sites
[0029] RNA extraction and reverse transcription: The lung tissues of Laiwu pigs and Dachang hybrid pigs were taken, and the total RNA of lung tissues was extracted according to the instructions of RNAsimple Total RNA Kit from TIANGEN Company. Then follow the TaKaRa PrimeScript TM RT reagent Kit with gDNA Eraser (Perfect Real Time) kit instruction manual for reverse transcription to obtain cDNA.
[0030] According to the mRNA sequence of SERPINA1 gene (NCBI Reference Sequence: NM_214395.1) design following primer SERPINA1CF gene sequence as shown in Seq ID No:1, SERPINA1CR gene sequence as shown in Seq ID No:2, it has included the SERPINA1 gene mRNA sequence The range from the 8th base to the 1273rd base (the sequence is shown in Seq ID No: 5).
[0031]
[0032] Use SERPINA1CF and SERPINA1CR primers to amplify the fragment from the 8th base to the 1273rd base of the SERPINA1 ...
Embodiment 2
[0034] Embodiment 2 ELISA detects the SERPINA1 serum content of different genotypes
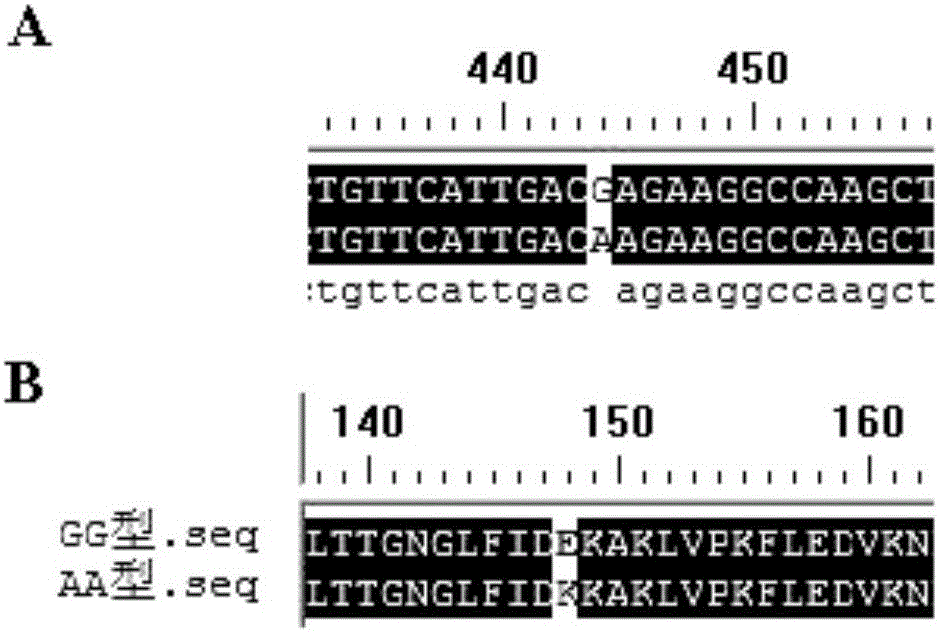
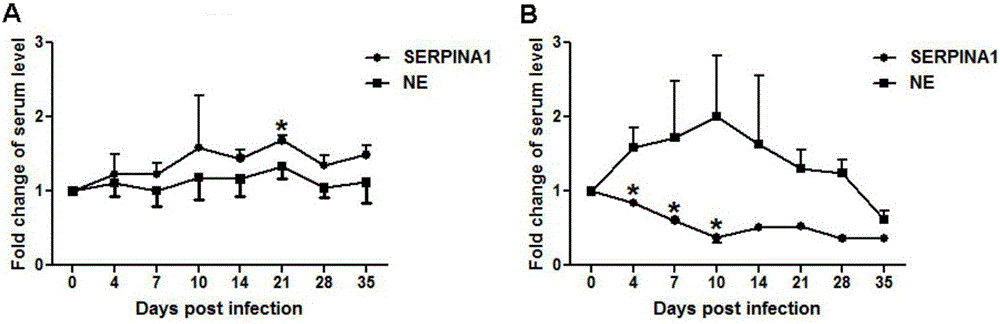
[0035] Take Laiwu pigs and Dachang hybrid pigs at 0, 4, 7, 10, 14, 21, 28 and 35 days after infection with PCV2 Serum, according to the porcine serine protease inhibitor 1 (SERPINA1 / A1AT) enzyme-linked immunosorbent assay kit Instructions for detecting serum levels of SERPINA1. Test results such as Figure 4 and Figure 5 Shown: In a healthy state, the serum level of AA-type individuals is the highest, followed by that of AG-type individuals, and the content of GG-type individuals is the lowest (P Figure 4 ). However, after infection with PCV2, the SERPINA1 serum level of GG individuals significantly increased at 4dpi (P Figure 5 ).
Embodiment 3
[0036] Example 3 Population Detection of Polymorphism at Locus 445 and Marker-Assisted Breeding
[0037] According to the gene sequence of SERPINA1 (NCBI Reference Sequence: NC_010449.4), the following primers were designed: the SERPINA1MF gene sequence is shown in Seq ID No: 3, and the SERPINA1MR gene sequence is shown in Seq ID No: 4. The range from the 4760th position to the 5629th position.
[0038]
[0039] Use SERPINA1MF and SERPINA1MR primers to amplify the fragment from position 4760 to position 5629 of the complete sequence of SERPINA1 gene. The amplification system is: 2×PrimeSTAR GC Buffer (Mg 2+ Plus) 10 μL, dNTPs 1.6 μL, each primer (10 μM) 0.5 μL, PrimeSTAR TM HS DNA Polymerase (5U / μL) 0.2μL, Genomic DNA 1μL, add ddH 2 O to a total volume of 25 μL. The PCR reaction conditions were 94°C for 5 min; 34 cycles of 98°C for 10 s, 56°C for 30 s, and 72°C for 1 min; 72°C for 8 min. The PCR product 870bp was obtained, and the amplified product was mixed with 6×Loa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com