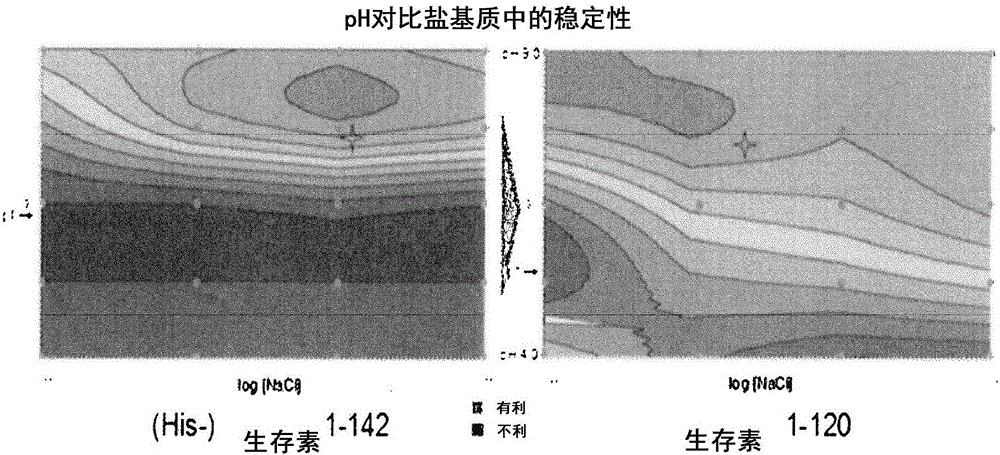
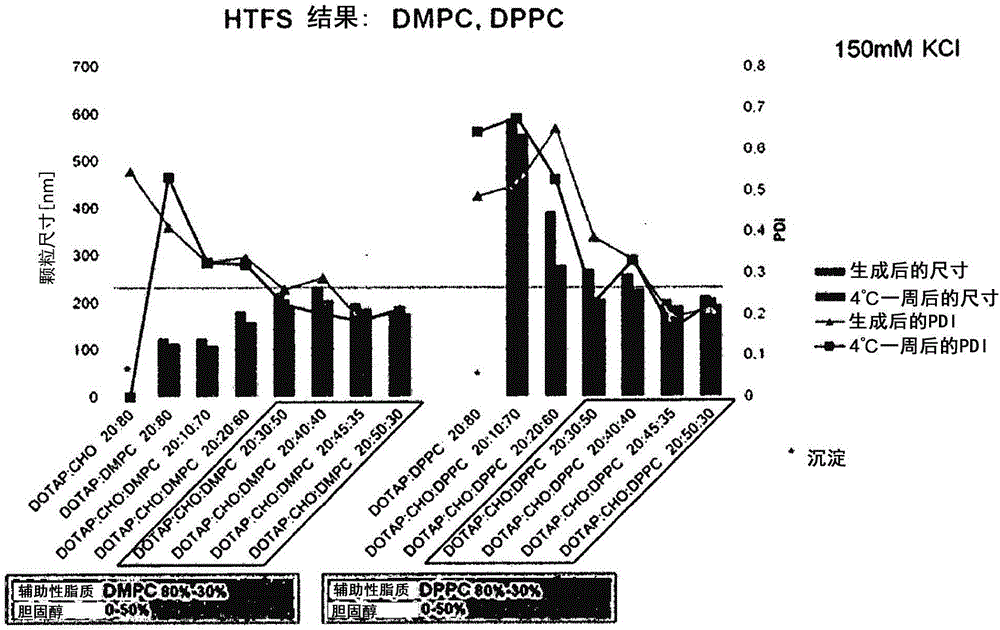
Survivin-directed cancer vaccine therapy
A survivin and vaccine technology, applied in the field of survivin-oriented cancer vaccine therapy, can solve the problems of insufficient immunogenicity, incompatibility, and reduced stability of survivin in vivo
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0234] Example 1: Material
[0235] The phospholipid substances used in the present invention are listed in Table 1. DOTAP was obtained from Merck Millipore and cholesterol was purchased from Sigma Aldrich. All other lipids were obtained from Lipoid AG. dC-survivin was prepared as described below and consisted of the amino acid sequence of human survivin truncated at the C-terminus for better stability characteristics and better manufacturability. It contains amino acids 2-120 of human survivin (SEQ ID NO: 3), and has the following characteristics: molecular weight 13.8 kDa, isoelectric point 6.0 and melting point 48°C.
Embodiment 2
[0236] Example 2: Engineering transformation of source strain Escherichia coli BL21 (DE3) and preparation of Escherichia coli T7E2 bacterial strain as the expression strain of the truncated survivin used in the present invention
[0237] The source strain Escherichia coli BL21 (DE3) (Novaki) was engineered as follows:
[0238] ·Incoming analysis of Escherichia coli BL21(DE3)
[0239] Changing the rpsL gene of E. coli BL21(DE3) genome to obtain streptomycin resistance phenotype
[0240] Substitution of non-essential DE3 prophage regions with the loxP-cmR–loxP cassette in the context of the E. coli BL21(DE3) STR genome
[0241] Replacement of Rac prophage with synthetic pgl gene in E. coli BL21(DE3)STR L'1DE3:loxP-cmR-loxP genetic background
[0242] Removal of loxP-flanked chloramphenicol markers
[0243] The genome sequence of E. coli BL21(DE3) [gb#CPOO 1509.3] and Artemis software (Rutherford et al., Bioinformatics 16(10):944-5, 2000) were used for in-silico strain engin...
Embodiment 3
[0245] Example 3: Transformation of Escherichia coli T7E with expression plasmid pAL37
[0246] The source plasmid was pET42 (Novaki), which was finally designed into the final expression plasmid pAL37 through pAL28. The plasmid DNA of pLA37 is shown entirely in Figure 20 and represented by SEQ ID NO:4. The restriction sites of the corresponding restriction enzymes are shown in Figure 19 . Plasmid AL37 contains DNA encoding the sequence of SEQ ID NO: 2 (1–120 aa of SEQ ID NO: 1) for a truncated survivin that is released into the medium after its expression, the first methionine residue The base was cleaved, thereby obtaining a representative truncated survivin SEQ ID NO:3 (2-120aa of SEQ ID NO:1).
[0247] For transformation, E. coli T7E2#1 cells were lysed on ice, 1 μl of plasmid al37 was added and incubated on ice for 30 minutes, at 42° C. for 30 seconds, and again on ice for 2 minutes. Then, add 250 μl of antibiotic-free LB Vegitone Broth and incubate at 37 °C for ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
| particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com