Method and kit for detecting free rare tumor cells in human biofluid sample
A technology of biological liquid and tumor cells, applied in the field of biological and medical detection, can solve the problems of loss of CTC, inability to accurately identify tumor cells, low expression of epithelial markers, etc., and achieve the effect of reducing the loss of CTC
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] Glucose analogue uptake assay in tumor cells
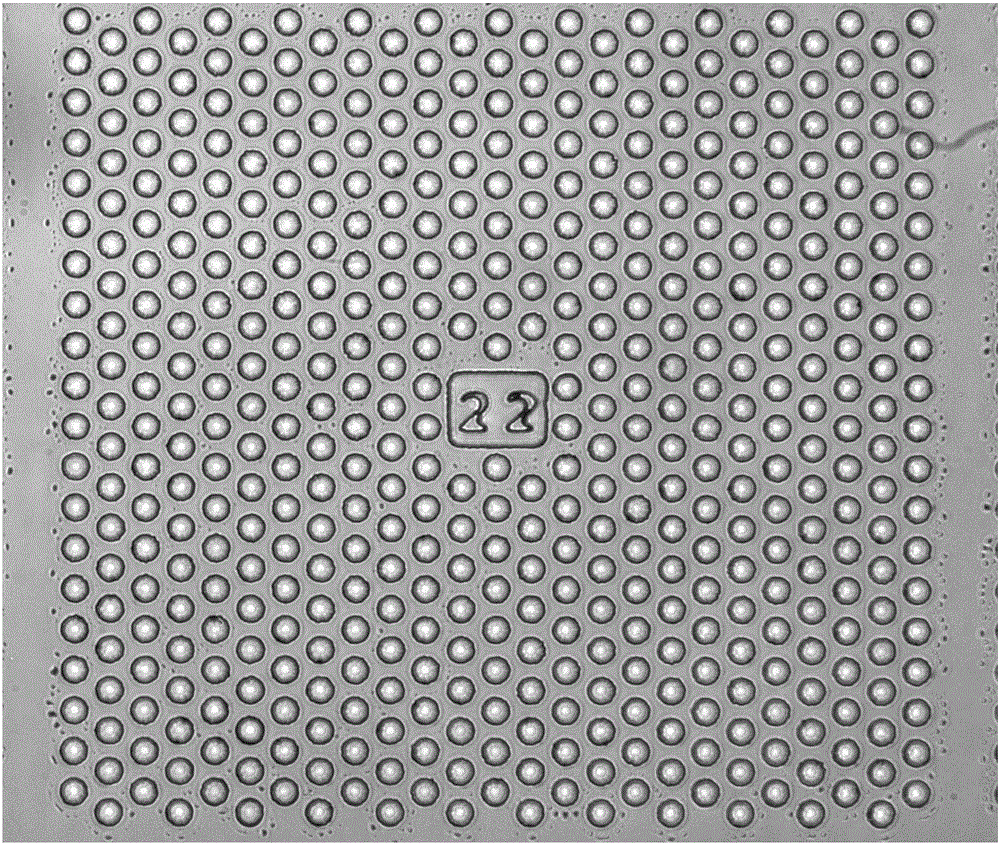
[0095] Provide a microwell array PDMS chip, its structure is as follows figure 1 As shown, each microwell is 20 microns in diameter and 30 microns deep, and every 500 microwells form a small area and are marked with numbers to realize the addressability of each microwell. A typical microwell chip consists of 400 such as Figure 1 The numerically coded small regions shown consist of 200,000 microwells.
[0096] Add the cell suspension of lung cancer cell H1650 on the microwell array chip, wash it with glucose-free cell culture medium RPMI-1640 after standing, as in figure 2 Cells are shown in most of the wells and at most one cell per well, with cells in the center of the well.
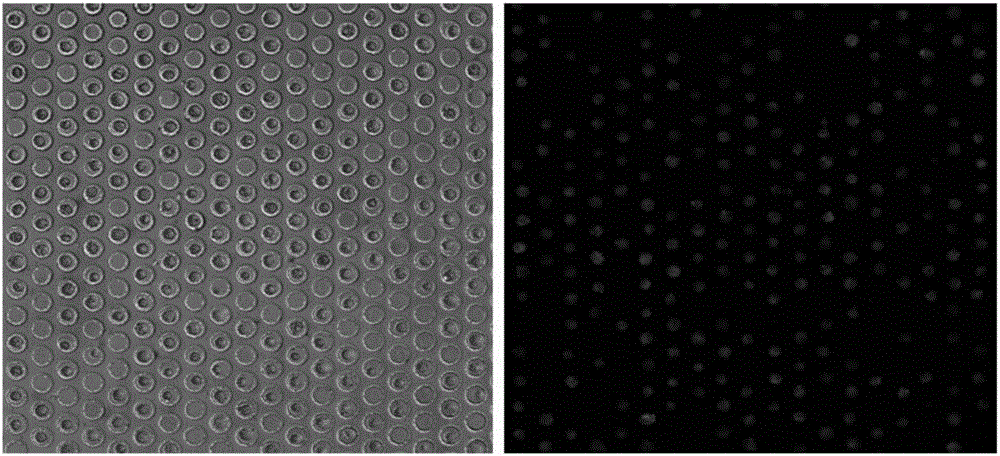
[0097] The tumor cells in the microwells were starved by incubating the cells with glucose-free cell culture medium RPMI-1640 for 10 minutes. Then, cell culture medium RPMI-1640 containing 2-NBDG (0.3 mM concentration) was added to incubate the...
Embodiment 2
[0099] Detection of Tumor Cells in Pleural Effusion Samples of Lung Cancer Patients
[0100] In this embodiment, the method includes the following steps:
[0101](1) After filtering 40 milliliters of pleural effusion of a lung cancer patient with 150 mesh gauze, centrifuge (500g, 5 minutes) to separate the cells, add 5 milliliters of red blood cell lysate (BD Company) to lyse in the dark for 5 minutes, and centrifuge again (500g, 5 minutes) ), after discarding the supernatant, resuspend and wash the cells with Hank's Balanced Salt Solution (HBSS), centrifuge (500g, 5 minutes), discard the supernatant, add 2 milliliters of HBSS to resuspend the cells;
[0102] (2) After cell counting, take 500 microliters of cell suspension (about 1 million cells), add 2 microliters of APC-labeled CD45 antibody, and incubate for 1 hour on an invertor;
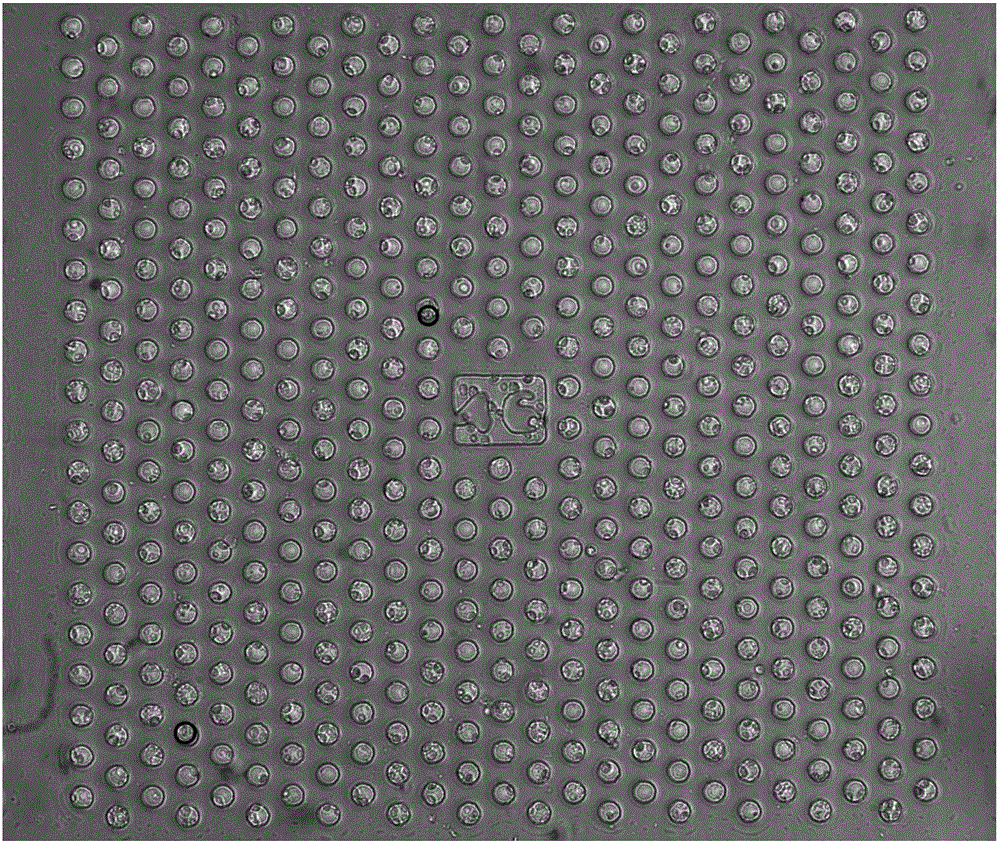
[0103] (3) Centrifuge, discard the supernatant and dilute the cells with HBSS, and drop the cell suspension on 2 microwell array chips (each c...
Embodiment 3
[0113] Detection of glucose analog uptake by tumor cells in peripheral blood samples of lung cancer patients
[0114] In this embodiment, the method includes the following steps:
[0115] (1) Take 1 ml of peripheral blood samples from patients with lung cancer, first centrifuge at low speed (200g, 5 minutes) to remove the platelet-rich plasma in the upper layer, resuspend the remaining cells in HBSS, add erythrocyte lysate (BD company) to lyse in the dark for 5 minutes, and again Centrifuge (500g, 5 minutes), discard the supernatant, resuspend and wash the cells with Hank's Balanced Salt Solution (HBSS), centrifuge (500g, 5 minutes), discard the supernatant, add 2 ml of HBSS to resuspend the cells;
[0116] (2) After counting cells, add CD45 antibody-labeled magnetic balls (Stemcell) according to the cell number 1:20, invert and incubate on an invertor, add 4 microliters of APC-labeled CD45 antibody after 15 minutes and continue inverting for 45 minutes;
[0117] (3) Transfer...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com