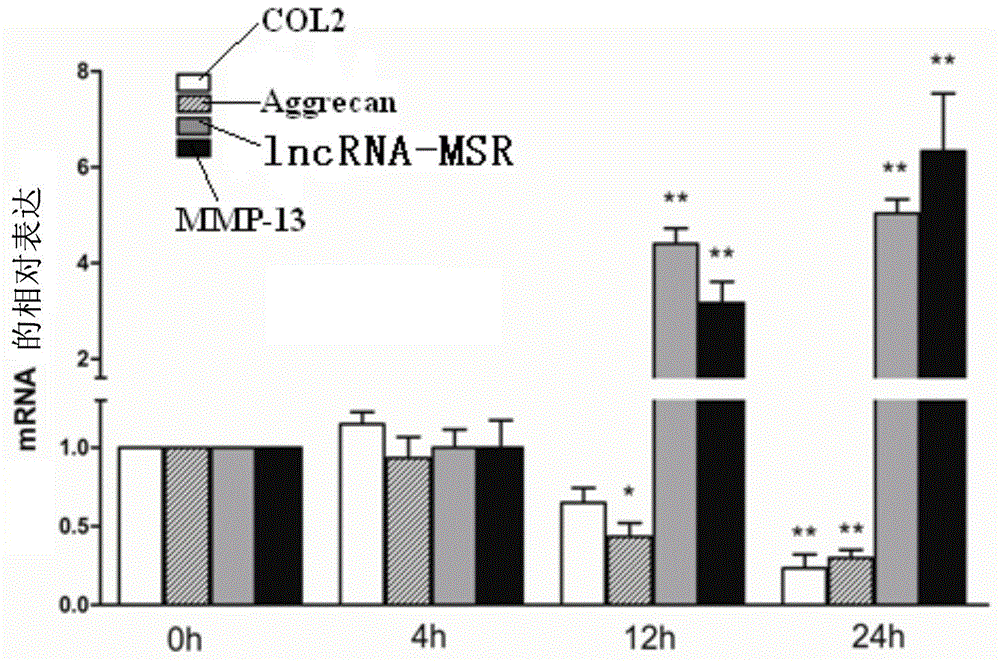
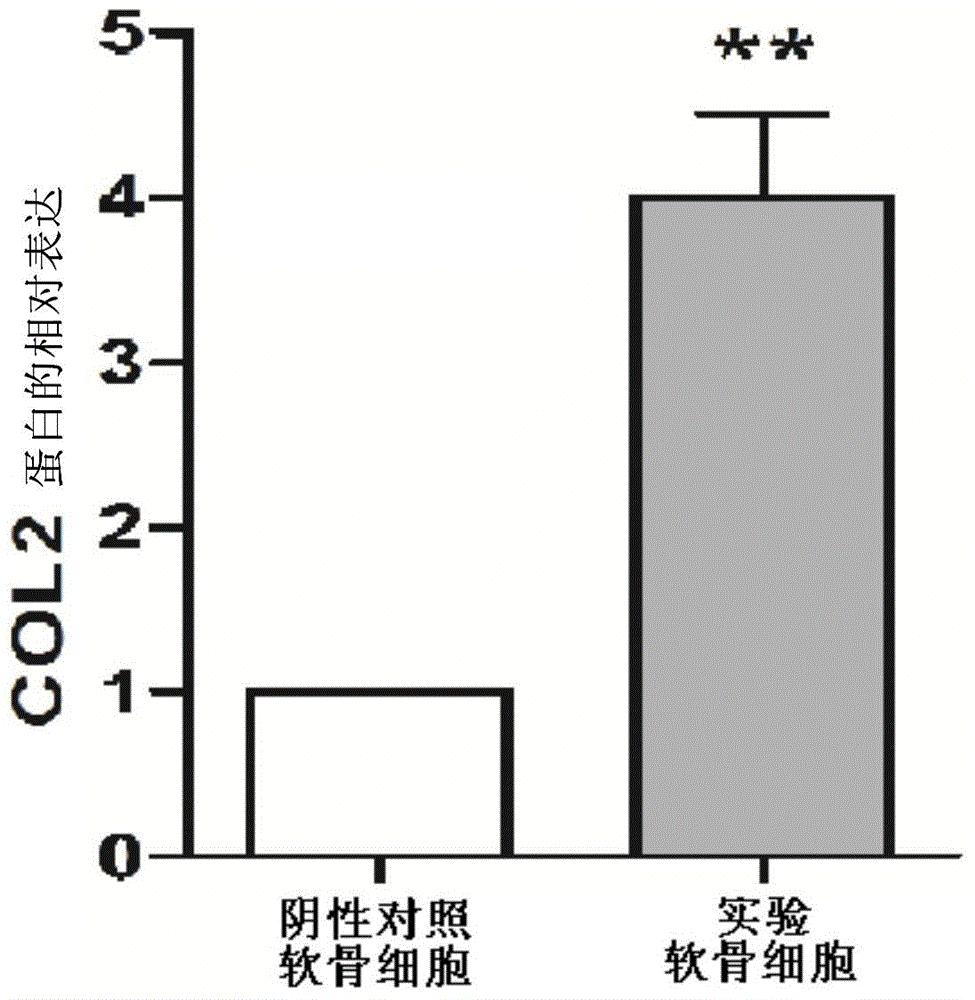
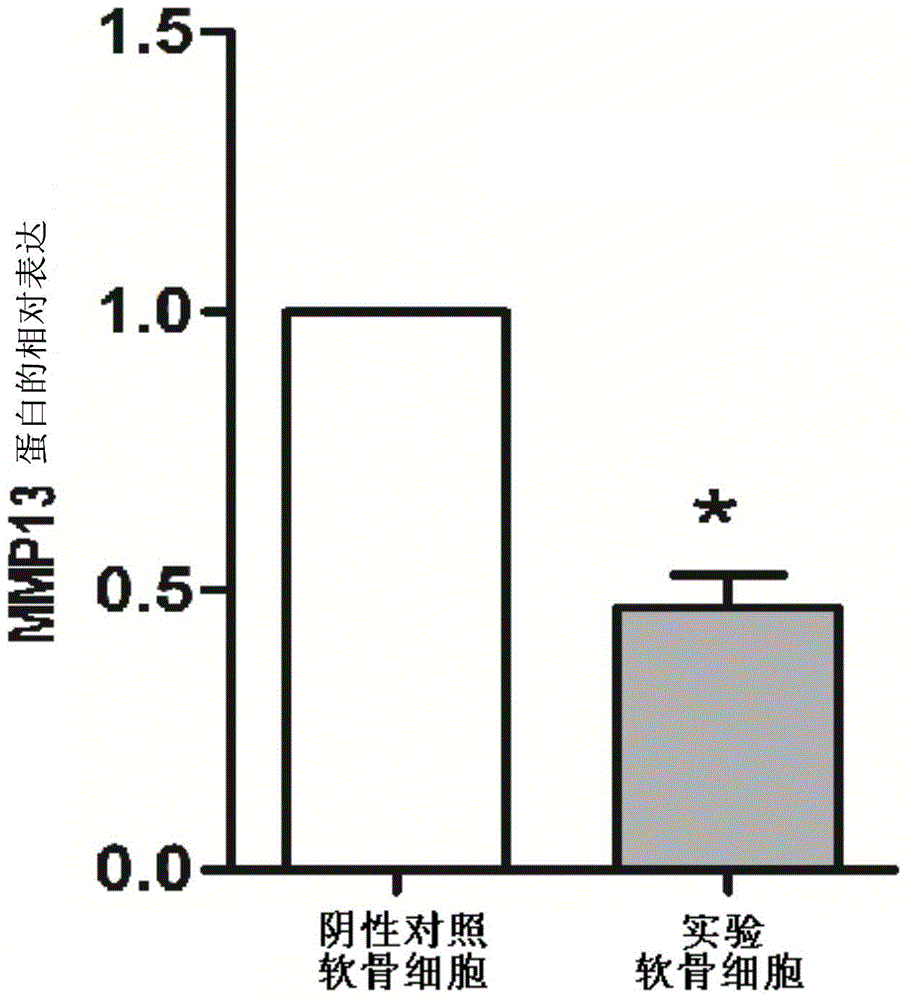
Inhibitor for lncRNA-MSR and application of inhibitor
An inhibitor and molecular technology, applied in the field of lncRNA-MSR inhibitors, can solve problems such as cartilage damage, cartilage tissue destruction, and level decline
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Step 1. Culture chondrocytes
[0062] Under sterile conditions, cut the cartilage tissue to less than 1cm with scissors 3 Size, and washed repeatedly with PBS buffer containing double antibody. Then, digest with trypsin in an amount 10 times the mass of cartilage tissue at 37° C. for half an hour, and discard the enzyme solution. Then utilize the PBS buffer solution of 0.2% (g / ml) type Ⅱ collagenase at the concentration of 10 times of the mass of cartilage tissue to continue to stir and digest at 37°C for 4 hours, and filter the treated cartilage after removing impurities. The tissue fragments are planted on the bottom surface with an area of 10cm 2 In a new petri dish, the culture medium is low-sugar DMEM complete medium, at 37°C, containing 5% volume concentration of CO 2 CO at saturated humidity 2 Constant temperature incubator culture.
[0063] Step 2, in vitro transfection of siRNA molecules
[0064] Negative control siRNA: The selected negative control siR...
Embodiment 2
[0107] The present embodiment utilizes Western Blot (Western Blot) to detect the expression levels of each protein in the experimental chondrocytes and negative control chondrocytes, and the specific steps are as follows:
[0108] Step 1. Prepare experimental chondrocytes and negative control chondrocytes, wherein the cultivation and transfection of the experimental chondrocytes and negative control chondrocytes are the same as those in Example 1.
[0109] Step 2, determination of total cell protein concentration and Western Blot experiment, wherein the protein extracted from cultured chondrocytes is used as a protein sample. The medium in the chondrocytes was washed twice with PBS buffer solution, and then 200 ul of protein lysate with a mass concentration of 2% SDS was added to each well of a six-well plate to obtain protein samples.
[0110] 2.1. Determination of total cell protein concentration
[0111] The preparation of bovine serum albumin BSA standard substance and th...
Embodiment 3
[0134] Step 1. Prepare experimental chondrocytes and negative control chondrocytes, wherein the cultivation and transfection of the experimental chondrocytes and negative control chondrocytes are the same as those in Example 1.
[0135] Step 2. Immunofluorescence staining of chondrocyte cytoskeleton
[0136] 2.1. At room temperature (about 25° C.), fix the cultured chondrocyte slide (cover glass) in a 4% paraformaldehyde solution for 10-15 minutes.
[0137] 2.2. Then rinse the chondrocytes with PBS buffer for 3 times, 5 minutes each time.
[0138] 2.3. Incubate the chondrocytes with 2% BSA solution at 37° C. for 30 minutes to block non-specific binding sites.
[0139] 2.4. Add the specific primary antibody diluted with 2% BSA solution (1:200 dilution), and then incubate overnight at 4°C.
[0140] 2.5. Add fluorescein-labeled secondary antibody (diluted at 1:200) diluted with 2% BSA solution, and incubate for 30 min in a wet box in the dark.
[0141] 2.6. Rinse the chondrocy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com