A kind of rna pulldown method and kit
A technology of RNA probes and magnetic beads, applied in biological testing, material inspection products, measuring devices, etc., can solve the problems of nuclease contamination, low binding specificity between RNA probes and proteins, false positives, etc., and achieve Prevent contamination, save experimental cost, and improve specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
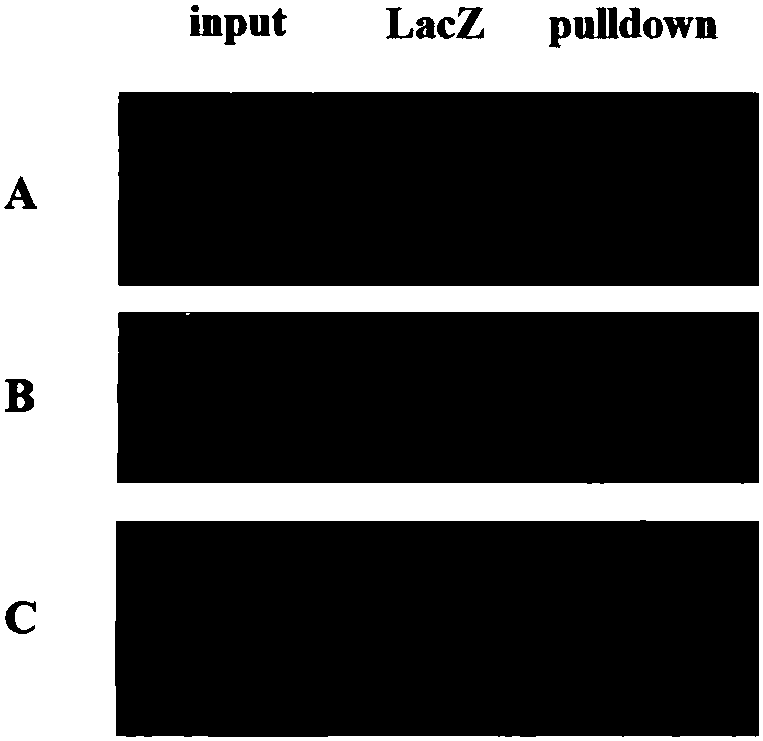
[0037] The present invention is illustrated by taking the pulldown effect of the lincRNA HOTAIR probe on the LSD1 protein as an example, and the probe is labeled with desthiobiotin 8-oxoG. The design and labeling methods of probes are well known to those skilled in the art and will not be repeated here. Streptavidin coupled to magnetic beads (BeaverBeads TM Streptavidin, Beaver Biotechnology Co., Ltd.) and affinity binding with dethiobiotin; the whole cell protein is incubated with magnetic beads-RNA probe, and the protein molecule can be specifically combined with the probe; after washing, the non-specific binding protein Molecule removal; finally, elution is performed with an eluent (for streptavidin) to obtain the target probe-protein complex, and then the protein type is identified by Western Blot. Specific steps are as follows:
[0038] S1. Magnetic bead pretreatment
[0039] ① Take 80 μL of streptavidin-containing magnetic beads and place them in an RNase-free EP tube...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com