Method for detecting miRNAs based on liquid chip of HCR
A DNA molecule and solution technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of lack of signal amplification system, inability to complete detection, etc., and achieve the effect of improving sensitivity, high sensitivity and good recovery rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0102] Example 1. Establishment of HCR-based miRNAs liquid-phase chip detection method
[0103] 1. Detection principle of miRNAs liquid chip based on HCR
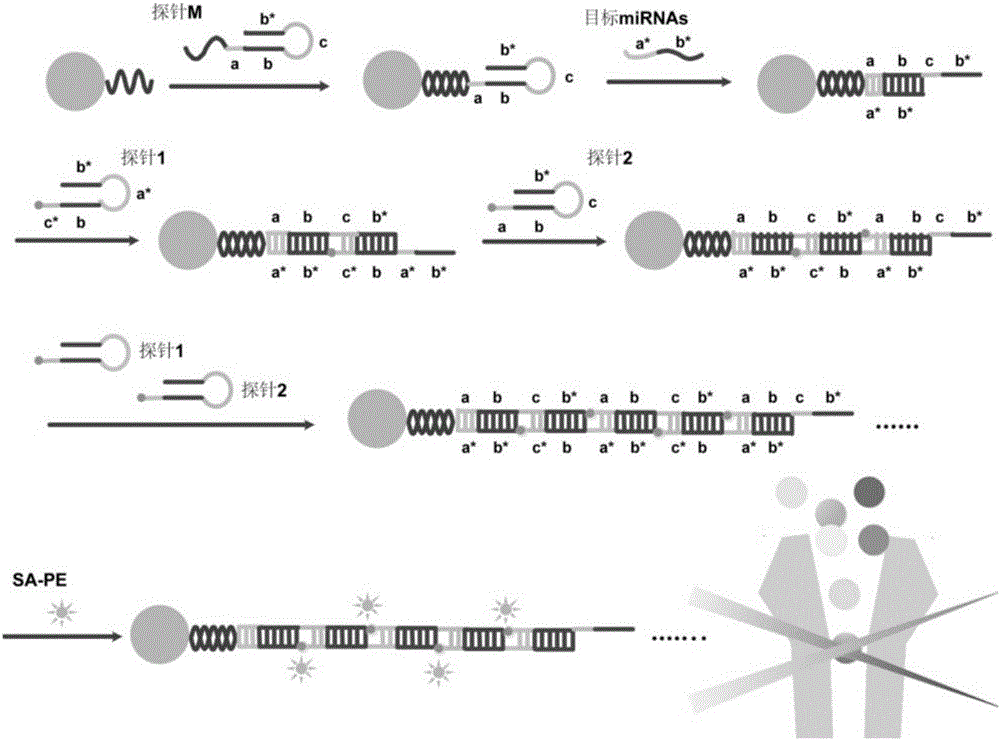
[0104] Schematic diagram of the principle of HCR-based miRNAs liquid chip detection figure 1 .
[0105] Probe M consists of 68 bases, including a sequence that is complementary to the capture sequence on the microsphere; a sequence that is complementary to the target miRNA (a-b); and a sequence that can trigger HCR to initiate a chain reaction (c-b*). After the probe M is denatured and annealed, a stem-loop secondary structure is formed, which is first detected in the system. -TAG TM Microsphere capture. When the target miRNA (a*-b*) is added, the miRNA hybridizes to the probe M, and the hairpin structure of the probe M is opened by a strand displacement reaction, exposing the HCR-triggered chain reaction (c-b* ).
[0106] Both probes 1 and 2 are composed of 44 bases, and are labeled with biotin at the 5' and 3' ends...
Embodiment 2
[0138] Example 2, HCR-based miRNAs liquid chip detection whether it contains let-7a
[0139] Use the miRBase database to query let-7a and miR-21 sequences:
[0140] let-7a: 5'-UGAGGUAGUAGGUUGUAUAGUU-3';
[0141] miR-21: 5'-UAGCUUAUCAGACUGAUGUUGA-3'.
[0142] Let-7a and miR-21 were artificially synthesized, diluted to 100 μM with TE buffer, and stored at -20°C to obtain let-7a standard solution and miRNA21 standard solution.
[0143] 1. Detection of let-7a content by HCR-based miRNAs liquid chip
[0144] 1. Selection of microspheres and design and synthesis of DNA hairpin probes
[0145] 1) Selection of microspheres
[0146] Buy size 12 and size 26 -TAG TM The microspheres were used to detect the content of let-7a and miR-21 respectively. Among them, No. 12 -TAG TM The cross-linked capture sequence on the microspheres is: 5'-AGTAGAAAGTTGAAATTGATTATG-3' (No. 12 microspheres are used to detect let-7a), No. 26 -TAG TM The capture sequence cross-linked by the microsph...
Embodiment 3
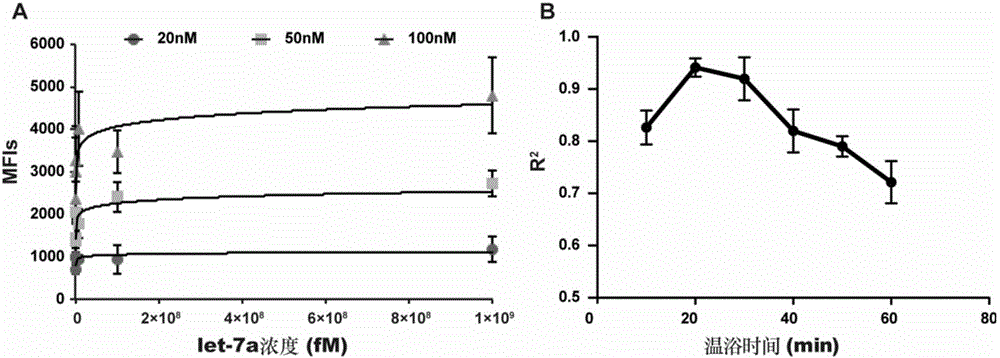
[0246] Embodiment 3, detecting target miRNA content in RNA to be tested
[0247] (1) Establish a standard curve
[0248] Dilute 100 μM let-7a standard solution to be tested with buffer A into 6 continuous concentration gradients of 0.1pM, 1pM, 10pM, 100pM, 1nM, 10nM as let-7a standard solution to be tested with different concentrations.
[0249] 1. Selection of microspheres and design and synthesis of DNA hairpin probes
[0250] 1) the selection of microspheres: the same as that of Example 2;
[0251] 2), design and synthesize corresponding probe M, probe 1 and probe 2 according to let-7a and miR-21: the same as that of Example 2;
[0252] 3) Probe pretreatment: the same as that of Example 2;
[0253] 2,
[0254] 1) The probe M is attached to the microsphere to obtain a microsphere-probe complex: the same as that of Example 2;
[0255] 2) The sample to be tested is hybridized with the microsphere-probe complex: the RNA solutions to be tested are respectively 0.1pM, 1pM, 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com