hUCMSCs (human umbilical cord mesenchymal stem cells) carrying PEDF (pigment epithelial-derived factor) and modified with recombinant lentivirus as well as preparation method of hUCMSCs
A technology of recombinant lentivirus and derivative factors, which is applied in growth factors/inducing factors, biochemical equipment and methods, cells modified by introducing foreign genetic materials, etc. Internal infection and other complications, to achieve the effect of low immunogenicity and no immune response
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
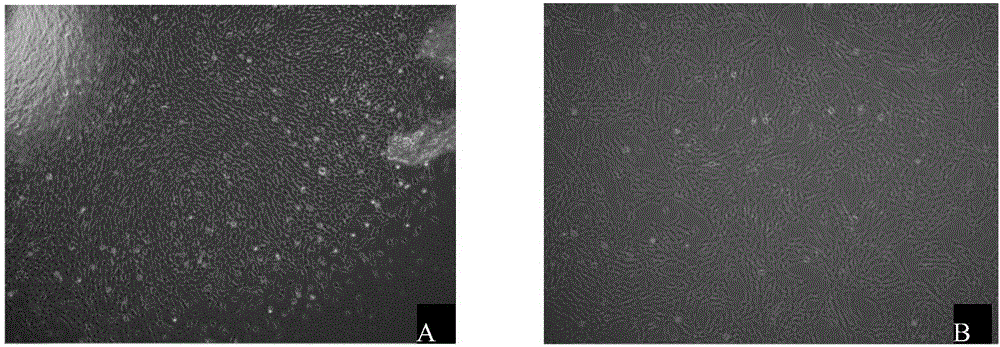
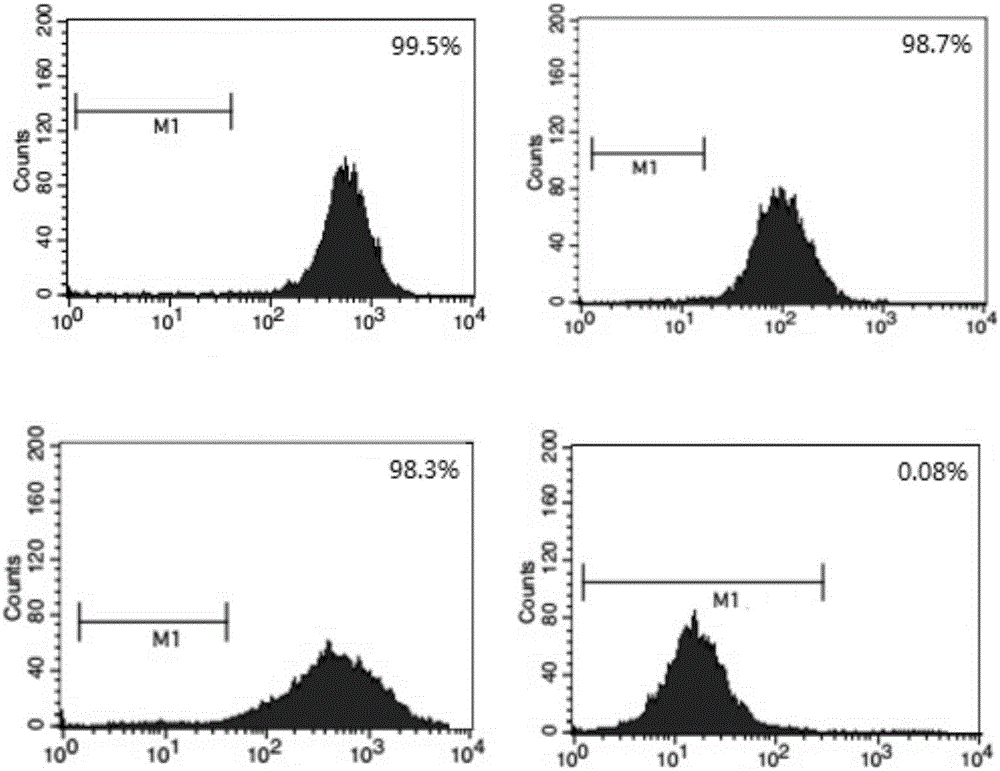
[0032] Isolation, culture and phenotypic identification of hUCMSCs.
[0033] Take the umbilical cords of normal full-term healthy newborns, cut Wharton’s jelly into 1mm×1mm×1mm size, digest with 0.1% collagenase and 0.125% trypsin for 30min, filter through 100 mesh and 200 mesh filters, and inoculate in containing 10% FBS in DMEM culture flask, put in 37℃, 5%CO 2 cultured in an incubator. Cell growth was observed daily with an inverted microscope. The primary culture can reach 90% growth and confluence after 12-15 days. After routine digestion with trypsin, 5000 cells / cm 2 Inoculation subculture expansion culture. When the passaged cells grow close to 80% confluence of the monolayer, they can continue to be passaged. The morphological characteristics of hUCMSCs were observed under an inverted microscope. The second generation hUCMSCs were used to detect the cell phenotype.
[0034] 24 hours after the primary inoculation, half the amount of medium was changed. It can be s...
Embodiment 2
[0037] Artificially synthesized PEDF gene.
[0038] Homo sapiens pigment epithelium-derived factor (PEDF) mRNA (AB593013.1) was retrieved from NCBI, and the 1257 bp of CDS region of PEDF gene is as follows:
[0039] 60a
[0040] 61 tgcaggccct ggtgctactc ctctgcattg gagccctcct cgggcacagc agctgccaga
[0041] 121 accctgccag ccccccggag gagggctccc cagaccccga cagcacaggg gcgctggtgg
[0042] 181 aggagggagga tcctttcttc aaagtccccg tgaacaagct ggcagcggct gtctccaact
[0043] 241 tcggctatga cctgtaccgg gtgcgatcca gcacgagccc cacgaccaac gtgctcctgt
[0044] 301 ctcctctcag tgtggccacg gccctctcgg ccctctcgct gggagcggag cagcgaacag
[0045] 361 aatccatcat tcaccggggct ctctactatg acttgatcag cagcccagac atccatggta
[0046] 421 cctataagga gctccttgac acggtcaccg ccccccagaa gaacctcaag agtgcctccc
[0047] 481 ggatcgtctt tgagaagaag ctgcgcataa aatccagctt tgtggcacct ctggaaaagt
[0048] 541 catatgggac caggccccaga gtcctgacgg gcaaccctcg cttggacctg caagagatca
[0049] 601 acaactgggt gcaggcgcag atgaaaggga agct...
Embodiment 3
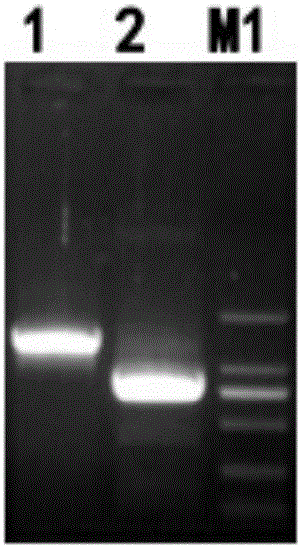
[0063] The PEDF gene fragment and the GFP fragment were amplified by PCR and cloned into the pLenti-CMV-hChR2-EYFP vector after fusion.
[0064] Using the T-Vector pMD19(Simple)-PEDF plasmid as a template, the CDS region of the PEDF gene was amplified by PCR, and the GFP sequence was amplified by PCR using the pLenti-CMV-hChR2-EYFP plasmid as a template. The reaction system is: T-Vector pMD19(Simple)-PEDF plasmid and pLenti-CMV-hChR2-EYFP plasmid (50-fold dilution) were 1 μL, 20 pmol / μL T-Vector pMD19(Simple)-PEDF plasmid primer 0.5 μL, 20 pmol / μL pLenti-CMV-hChR2-EYFP plasmid primer 0.5 μL, PrimeSTAR HS (Premix) 25 μL, add dHO 2 O to a total volume of 50 µL. Reaction conditions: 98°C for 10s, 55°C for 15s, 72°C for 1min, 30 cycles. Obtain PCR products A and B, take 5 μL for 1% agarose gel electrophoresis. Use Takara MiniBEST Agarose Gel DNA Extraction Kit Ver.4.0 to cut the gel and recover the target fragments, which are named PEDF fragments and GFP fragments respectively....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com