Maize transcription factor zmbhlh2 and its application
A technology of transcription factor and maize, applied in the field of genetic engineering, can solve the problem that the function of maize bHLH transcription factor is poorly understood, and achieve the effects of improving plant drought resistance, improving tolerance and small environmental impact
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
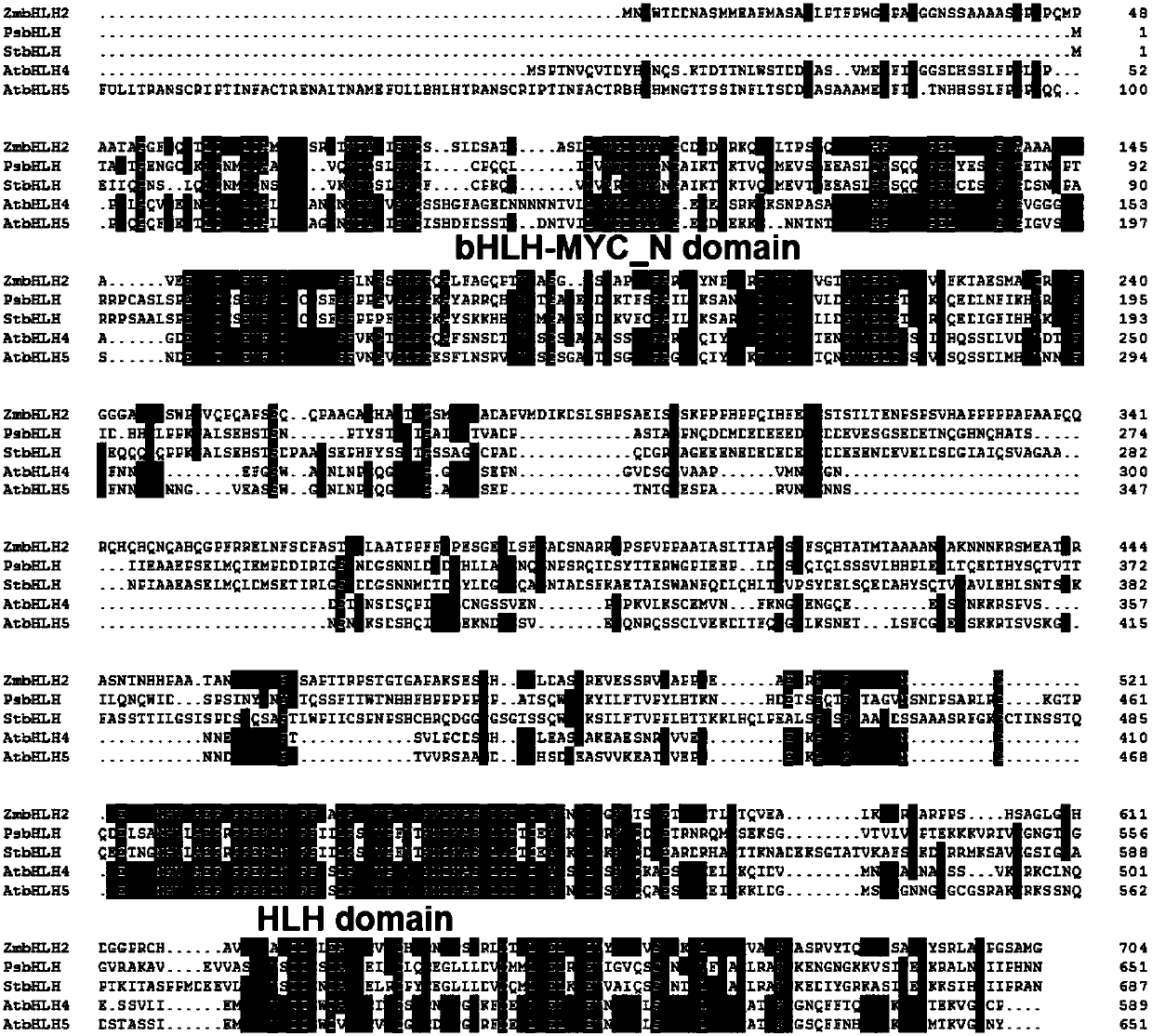
[0039] Cloning of Example 1 Maize Transcription Factor ZmbHLH2 Gene
[0040] The maize transcription factor ZmbHLH2 gene was cloned from the leaves of the maize variety Dongnong 255 (this maize variety has been approved and promoted by relevant departments) cultivated by the College of Agriculture of Northeast Agricultural University by RT-PCR technology. The specific method is as follows:
[0041] 1. Extract the total RNA from the maize leaves, and reverse transcribe to obtain the first strand of cDNA.
[0042] 2. The cDNA sequence of the transcription factor ZmbHLH2 gene was obtained by PCR amplification reaction using the first strand of DNA as a template and ZmbHLH2F and ZmbHLH2R as primers.
[0043] Wherein, the sequences of primers ZmbHLH2F and ZmbHLH2R described in step (2) are as follows:
[0044] ZmbHLH2F: 5′-ATCCGCATTCTGAACCATT-3′
[0045] ZmbHLH2R: 5'-TGCCTATCGTGTTGTAACC-3'.
[0046] Use 2×Taq PCR MasterMix (Tiangen Biochemical) for PCR amplification, the reactio...
Embodiment 2
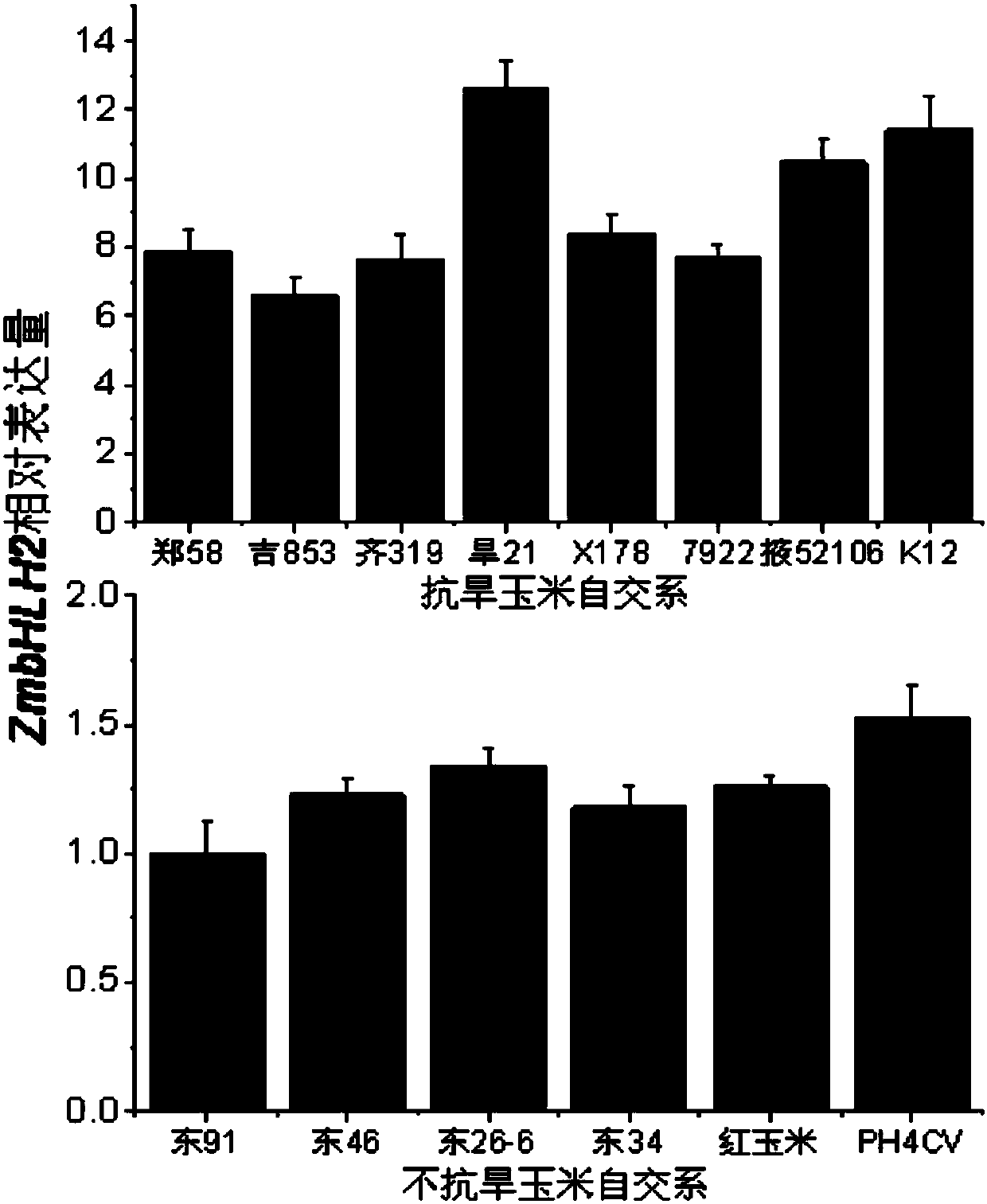
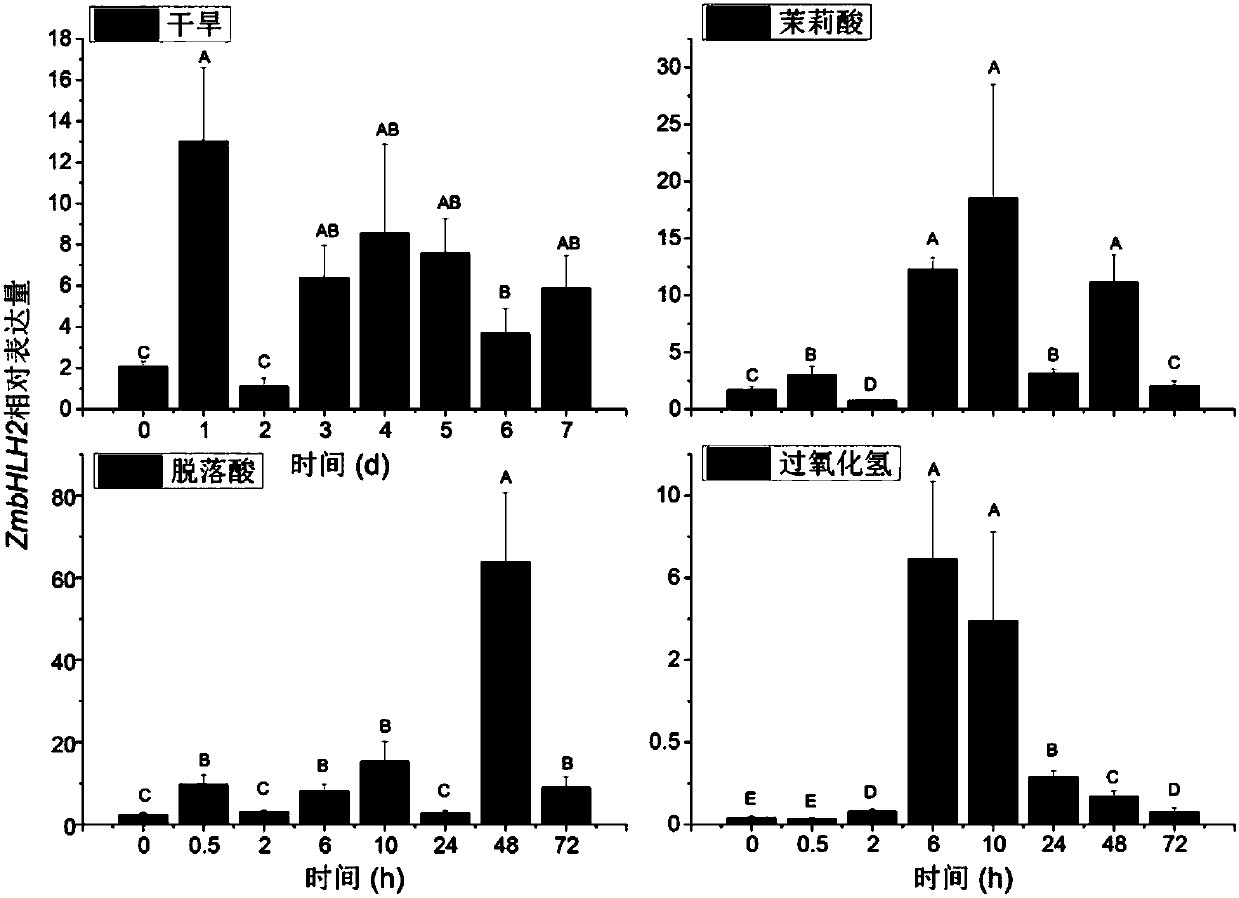
[0049] Expression analysis of embodiment 2 ZmbHLH2 gene when subjected to stresses such as drought
[0050] The expression pattern of ZmbHLH2 gene in different drought-resistant inbred lines was detected by fluorescent quantitative PCR. Drought-resistant inbred lines, Zheng 58, Ji 853, Qi 319, Han 21, X178, 7922, Ye 52106 and K12; drought-sensitive inbred lines, Dong 91, Dong 46, Dong 26-6, Dong 34, red corn and PH4CV (See figure 2 ), the expression of ZmbHLH in drought-resistant inbred lines was significantly higher than that in non-drought-resistant inbred lines.
[0051] The following fluorescent quantitative PCR detection primers were designed according to the ZmbHLH2 gene sequence:
[0052] ZmbHLH2-QF1: 5′-GACGCCATCCTCCTACATC-3′
[0053] ZmbHLH2-QR1: 5′-GCTCTCCTCCTTGATTACC-3′
[0054] ZmACTIN-F: 5′-ACCTCACCGACCACCTAATG-3′
[0055] ZmACTIN-R: 5′-GCAGTCTCCAGCTCCTGTTC-3′
[0056] ZmACTIN (GenBank accession number: GRMZM2G126010) was used as an internal reference gene....
Embodiment 3
[0058] Example 3 Obtaining and Drought Resistance Verification of ZmbHLH2 Gene-transferred Arabidopsis Plants
[0059] Total RNA was extracted from maize leaves, and the first strand of cDNA was obtained by reverse transcription. The cDNA sequence of the transcription factor ZmbHLH2 gene was obtained by PCR amplification reaction using the first strand of DNA as a template and ZmbHLH2F and ZmbHLH2R as primers, and its cDNA fragment was ligated into the pMD18-T vector and transformed into Escherichia coli DH5α strain for cloning and Sequencing verification, the correct plasmid was named ZmbHLH2-18T; using the ZmbHLH2-18T plasmid as a template, ZmbHLH2-ORF-F: 5′-CG GAATTC ATGAACCTGTGGACG-3' (the underline is the EcoR I restriction site) and ZmbHLH2-ORF-R:5'-CG GGATCC CCTGCCCATGGCAGA-3' (the underline is the BamHI restriction site) is used as a primer to amplify the ORF of the ZmbHLH2 gene, and the amplified product is double-digested by EcoR I and BamHI and then connected to t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com