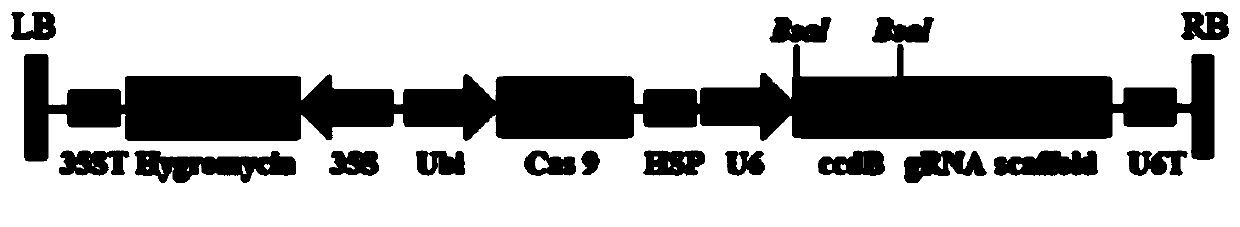A method for targeted knockout of rice miRNA
A rice and carrier technology, applied in the field of plant genetic engineering, can solve the problems of large blindness, difficulty in obtaining mutants, time-consuming and labor-intensive problems, and achieve the effect of simple operation and high knockout rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
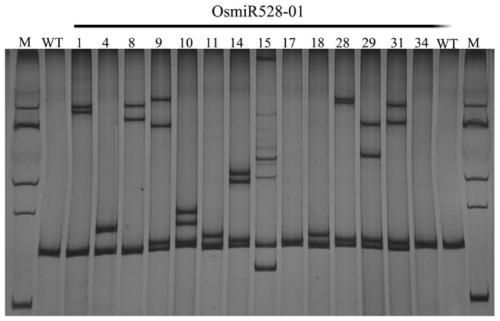
[0039] Example 1 Using the method of the present invention to create a single miRNA mutant (taking rice OsmiR528 as an example)
[0040] 1. Vector construction of CRISPR-Cas9-miRNA mutant
[0041] (1) Primer design
[0042] The primers are designed according to the CRISPR / Cas9 recognition and cutting rules of the target site. For the precursor genome sequence of rice OsamiR528 (GenBank number: GQ419957.2), the primers were designed as OsmiR528-sgRNA1-F: 5'-gtgtGAAGGGGCATGCAGAGGAGC-3'; OsmiR528-sgRNA1-R: 5'-aaacGCTCCTCTGCA TGCCCCTTC-3'.
[0043] (2) Primer annealing
[0044] Dilute the upstream and downstream primers of each target site by 10 times, take 10 μL of each, at 98° C., denature for 5 minutes, cool naturally, and dilute the annealed product by 20 times for use.
[0045] (3) Enzyme digestion, glue recovery, and connection
[0046] The backbone vector used in the experiment is pZHY988 ( figure 1 ), constructed by our laboratory, the construction process is: first synthesis (by Sha...
Embodiment 2
[0090] Example 2 Using the method of the present invention to create multiple miRNA mutants (taking rice OsmiR397a and OsmiR97b as examples)
[0091] 1. Vector construction of CRISPR-Cas9-miRNAs mutant
[0092] (1) OsmiR397a and OsmiR397b double-site knockout vector construction method
[0093] The OsmiR397 family has two members, OsmiR397a (GenBank number: AP014962:28489785...28489898) and OsmiR97b (GenBank number: XM_015769221). In order to obtain the OsmiR397 knockout mutant, a two-site knockout mutation vector was constructed in which both OsmiR397a and OsmiR97b were knocked out simultaneously. According to the precursor sequences of OsmiR397a and OsmiR97b, design and synthesize OsmiR397a-sgRNA1, gRNA scaffold, U6 terminator, U6 promoter, OsmiR397b-sgRNA1 sequence OsmiR397a-gRNA1-OsmiR397b-gRNA1-F and OsmiR397a-gRNA1-OsmiR397a-gRNA1-Osmi R (synthesized by Shanghai Yingjun Biotechnology Co., Ltd.). OsmiR397a-gRNA1-OsmiR397b-gRNA1-F:
[0094] gtgtGAGTGCAGCGTTGATGAACAAGTTTTAGAGCTA...
Embodiment 3
[0105] Example 3 Using the method of the present invention to create a large-fragment deletion mutant of miRNA (taking rice OsmiR408 as an example)
[0106] 1. Vector construction of CRISPR-Cas9-miRNAs mutant
[0107] (1) OsmiR408-sgRNA1 and OsmiR408-sgRNA2 double-site knockout vector construction method
[0108] When considering knocking out all or most of the precursor miRNA or it is difficult to find a suitable PAM site near certain mature miRNA precursors, it is necessary to consider the strategy of using double-site knockout to create large fragment deletions.
[0109] According to the precursor sequence of OsmiR408, design and synthesize the sequence OsmiR408-gRNA1-gRNA2-F and OsmiR408-gRNA1-gRNA2-R containing sgRNA1, gRNA scaffold, U6 terminator, U6 promoter, and sgRNA2 (by Shanghai Yingjun Biotechnology) Co., Ltd. synthesis).
[0110] OsmiR408-gRNA1-gRNA2-F:
[0111] gtgtGATGAGGCAGAGCATGGGATGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTTCTAGA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com