Amplicon primer and establishment method for establishing variable region sequencing library of microorganism bacterium 16s rDNA
A technology for sequencing libraries and sequencing primers, applied in biochemical equipment and methods, microbial determination/inspection, DNA preparation, etc., can solve the problem of increasing 16S rDNA sequencing steps and costs, the inability of sequencing systems to correct data, and increasing the complexity of sequencing libraries, etc. problem, achieve the effect of good lysing bacteria, increase the complexity, simplify the process steps and time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
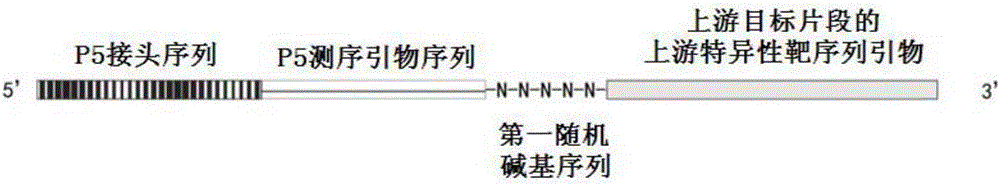
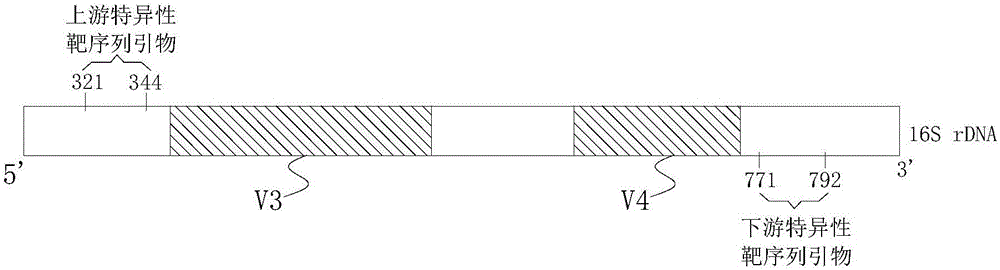
[0044] A primer for constructing a microbiota bacterial group 16s rDNA amplicon library, the primer is composed of an upstream primer and a downstream primer, the upstream primer includes an upstream specific target sequence primer, and the primer length of the upstream specific target sequence primer is 24nt, The primer sequence is: SEQ ID NO.1: 5'-CCANACTCCTACGGGAGGCAGCAG-3', the fourth base N in this SEQ ID NO.1 is A, C, G or T, which can be selected from A, C, G and any one of T. A first random base sequence is connected to the 5' end of the upstream specific target sequence primer. The first random base sequence is a sequence consisting of 5 random bases, and each random base is A, C, G or T, that is, each random base is selected from any one of A, C, G and T. In addition, a P5 sequencing primer sequence is connected to the 5' end of the first random base sequence, and a P5 linker sequence is also connected to the 5' end of the P5 sequencing primer sequence.
[0045] Th...
Embodiment 2
[0057] A method for constructing the 16s rDNA amplicon library of the microbiota bacterial group, specifically as follows, see also Figure 4 :
[0058] S1: Prepare upstream primers and downstream primers, refer to Example 1 for details, and do not repeat the limitations here;
[0059] S2: extracting microbial bacterial DNA in the sample;
[0060] S3: Take 10-50ng of extracted microbial bacterial DNA (total volume 2 0 to a total volume of 20ul; pipette the uniform system; put the sample tube on the PCR amplification instrument, set the program as a pre-denaturation temperature of 96 ° C for 2 minutes, and then enter the TouchDown PCR (falling PCR) step for the first round of amplification The denaturation temperature is 96°C and heated for 30s, the first cycle annealing temperature is 70°C, the annealing time is 1min, the extension temperature is 72°C, and the extension time is 2min, and then the annealing temperature is decreased by 0.7°C in each cycle, and a total of 20 cyc...
Embodiment 3
[0065] Experimental group: the amplified product obtained by TouchDown PCR in Example 2.
[0066] Control group: using the ordinary PCR amplification method, the samples, primers, reagents, and the sample volume of various components are consistent with the system in Example 2, and the ordinary PCR amplification procedure is used, as follows: the first step of pre-denaturation temperature 96 Heating at ℃ for 2min, heating at 96℃ for 30s in the second step, annealing temperature in the third step at 56℃, annealing time for 1min, extension temperature in the fourth step at 72℃, and extension time for 2min, followed by 34 cycles from the second step to the fourth step Cycle, after a total of 35 cycles, set at 72°C for 2 minutes, and finally lower the temperature to 4°C for storage.
[0067]The expected size of the amplified product in both the experimental group and the control group was 580bp. 5 ul of the amplification products obtained in the experimental group and the control...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com