Histone acetylation modified MT2A genome detection kit and primer pairs
A detection kit and primer pair technology, applied in the field of genome detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Example 1: Design of special primers for MT2A genomic DNA histone acetylation
[0086]According to the distribution characteristics of histone modification in the genome, the specific MT2A gene sequence was selected from GenBank, and the DNAMAN 6.0.40 software was used for comparison and analysis to find out the conserved gene sequence of MT2A, and the primers were designed with the software Primer Express 3.0, and the upstream primer MT2A-FP : the nucleotide sequence is: 5'-AGGTTATTCGGAGCCCCCTT-3', (SEQ ID No.1), the downstream primer MT2A-RP: the nucleotide sequence is: 5'-GTTCCTCAGTCCACAACCGA-3', (SEQ ID No.2) ;
[0087] All primers were synthesized by Tianyi Huiyuan Technology Co., Ltd.
Embodiment 2
[0088] Example 2: The composition of the detection kit for MT2A genomic DNA histone acetylation
[0089] MT2A genomic DNA histone acetylation detection kit, which is shown in the primer pair shown in the sequence table SEQ ID No.1 to SEQ ID No.2, as shown in the sequence table SEQ ID No.3 to SEQ ID No.4 Positive control primer pair, chromatin immunoprecipitation reagent set, nucleic acid extraction reagent and PCR reaction reagent.
[0090] The primers used to detect histone acetylation of MT2A genomic DNA are shown in Example 1;
[0091] 1% formalin, PBS, lysis buffer, dilution buffer, TSE I, TSE II, Buffer III, TE buffer, elution buffer of the chromatin immunoprecipitation reagent group, the preparation methods of the above reagents as follows:
[0092] Dilute 1% formalin: 37% formaldehyde with PBS at a volume ratio of 1:36.
[0093] Lysis buffer: 1% SDS, 5mM EDTA, 50mM Tris.HCl[pH 8.0], protease inhibitor cocktail;
[0094] Dilution buffer: 1% Triton X-100, 2mM EDTA, 15...
Embodiment 3
[0103] Example 3: Specificity test of MT2A genomic DNA histone acetylation
[0104] 1. Preparation of samples
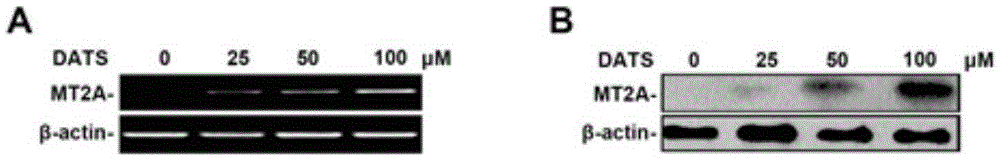
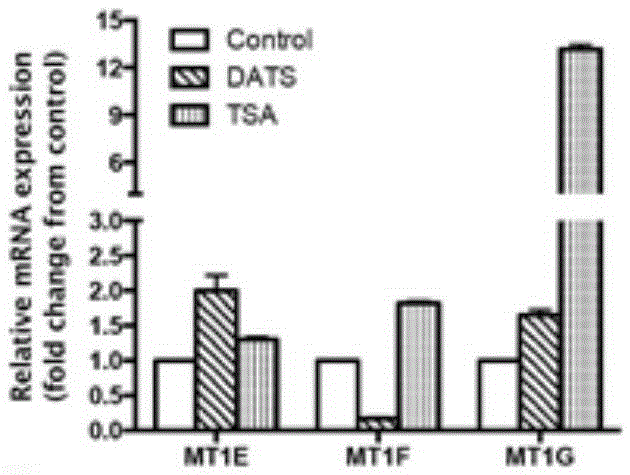
[0105] Gastric cancer cells were inoculated in a 10cm-diameter culture dish, and when the cells grew to about 60-80%, DATS (40 μM) was added for 12 hours, normal saline was added to the negative control group, and TSA (5 nM) was added to the positive control group for 24 hours. Collect 3-5 Petri dishes per group.
[0106] 2. Collect and wash samples
[0107] The cells were washed 1-2 times with room temperature pre-warmed PBS, and then treated with 1% formalin at 37°C for 10 min. Wash the cells twice with ice-cold PBS, then transfer to 1ml of ice-cold PBS with a cell scraper, centrifuge at 3,000rpm at 4°C for 2min, and discard the supernatant. Resuspend the cell pellet with 400 μl lysis buffer and incubate on ice for 10 min.
[0108] 3. Ultrasonic fragmentation of chromatin DNA, sonication 16 times, 2s each time, centrifugation at 4°C, 14,000g for 15min, and coll...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com