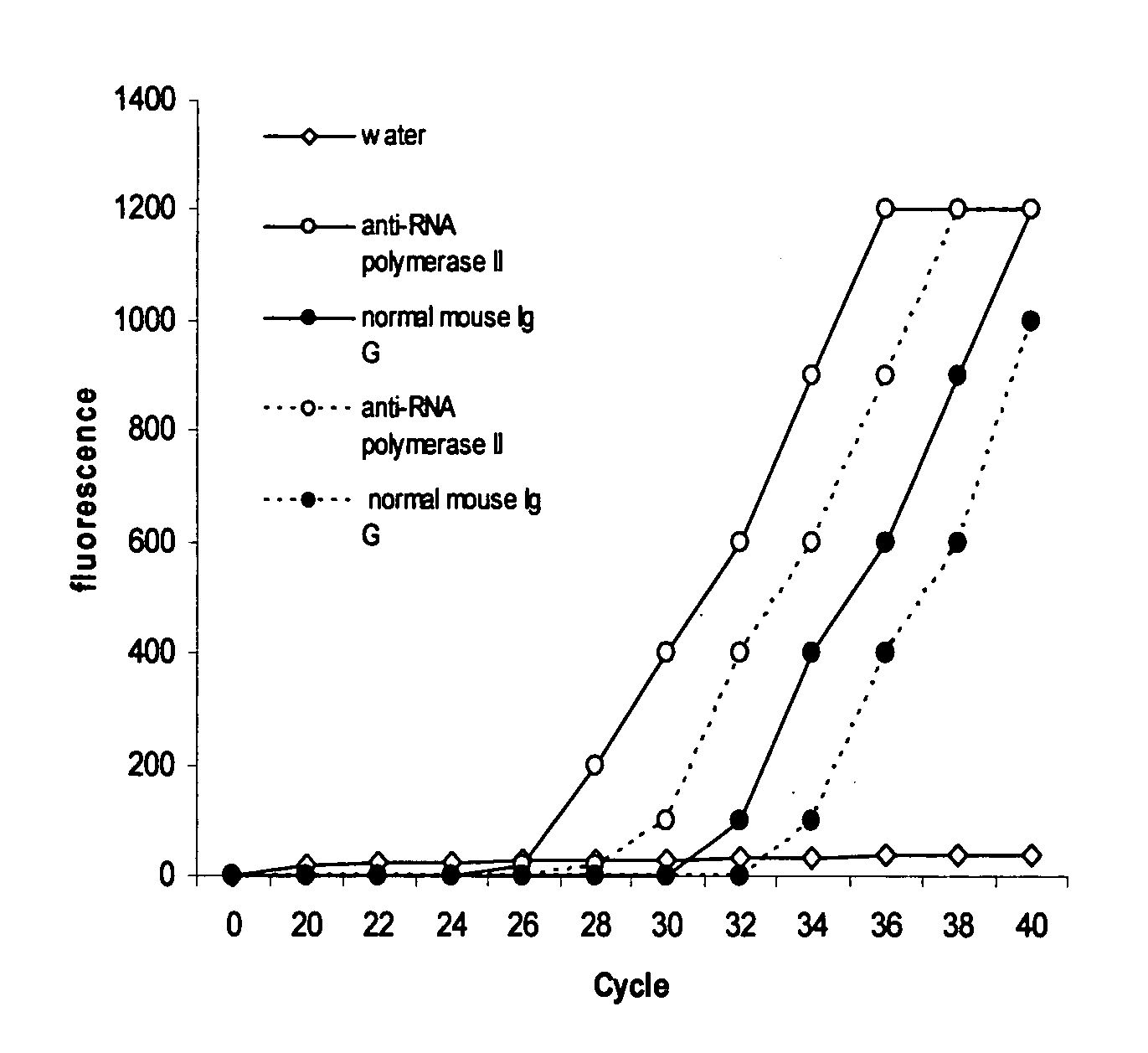
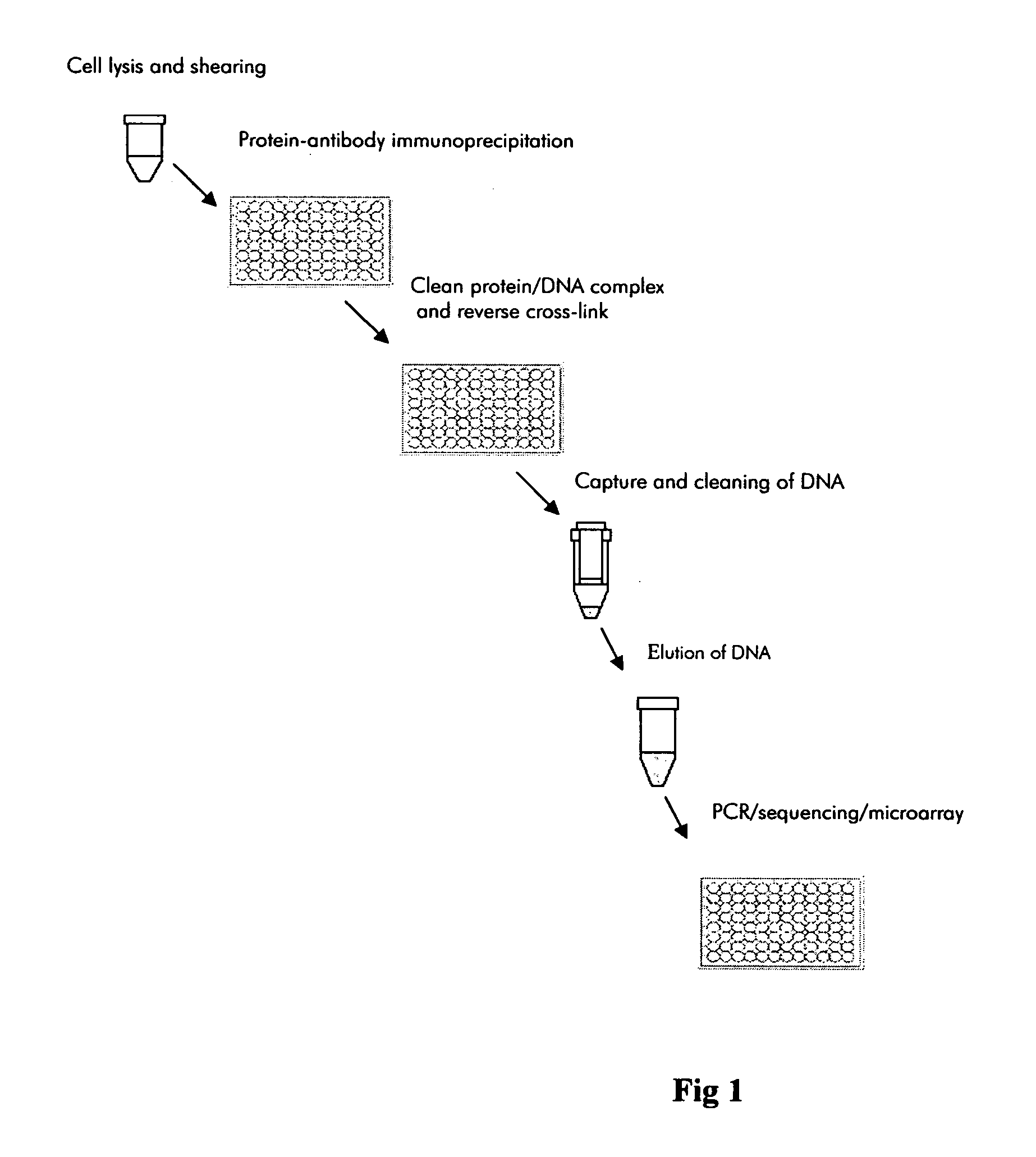
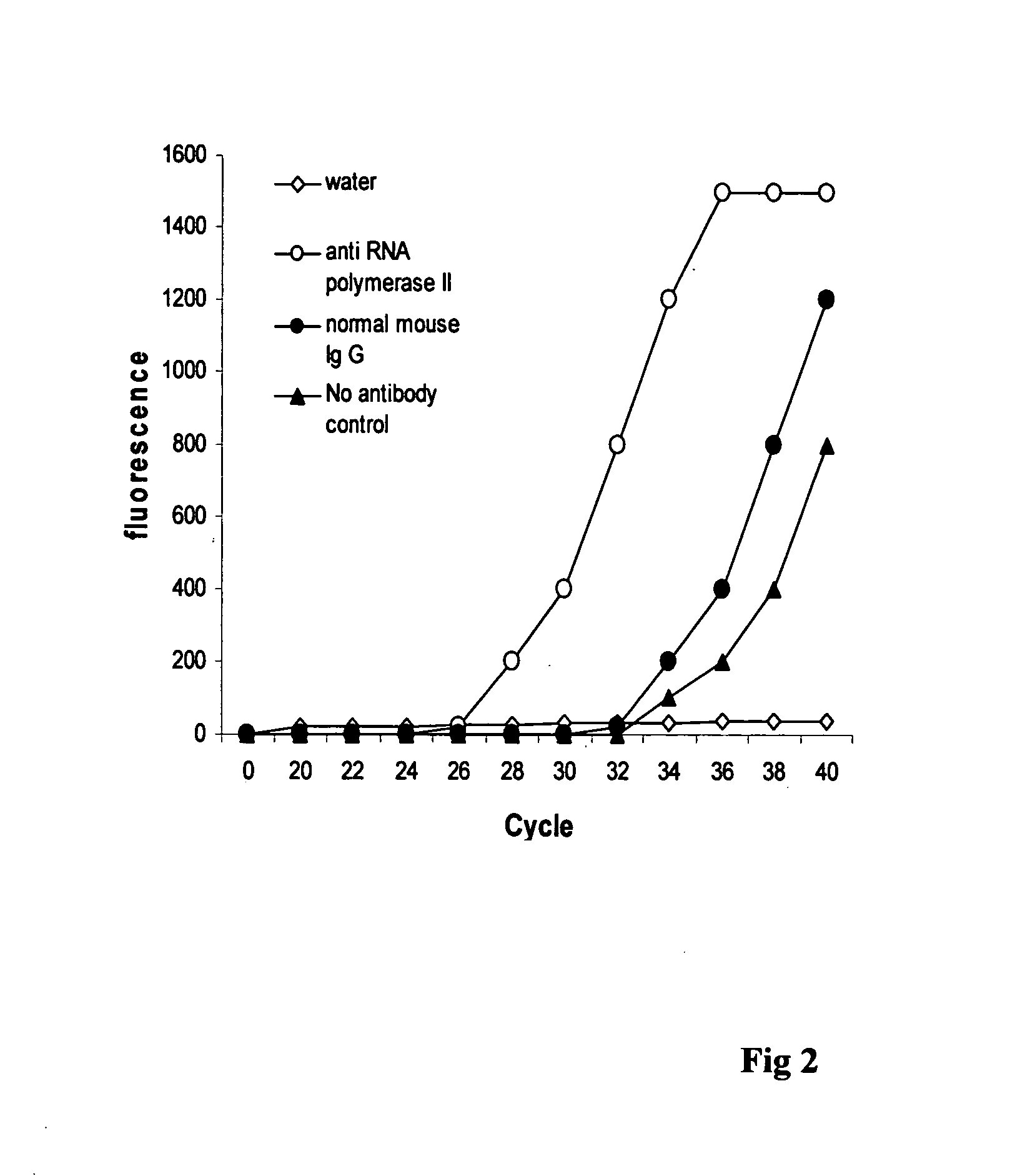
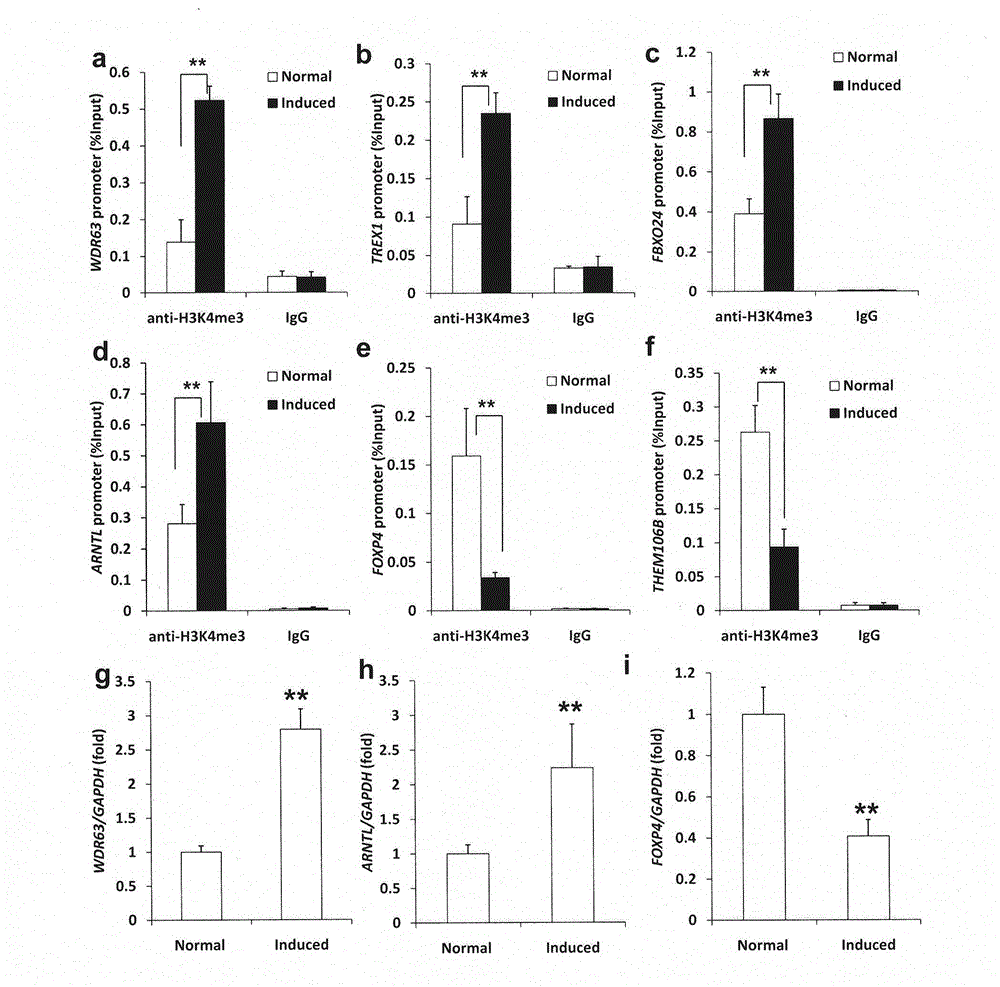
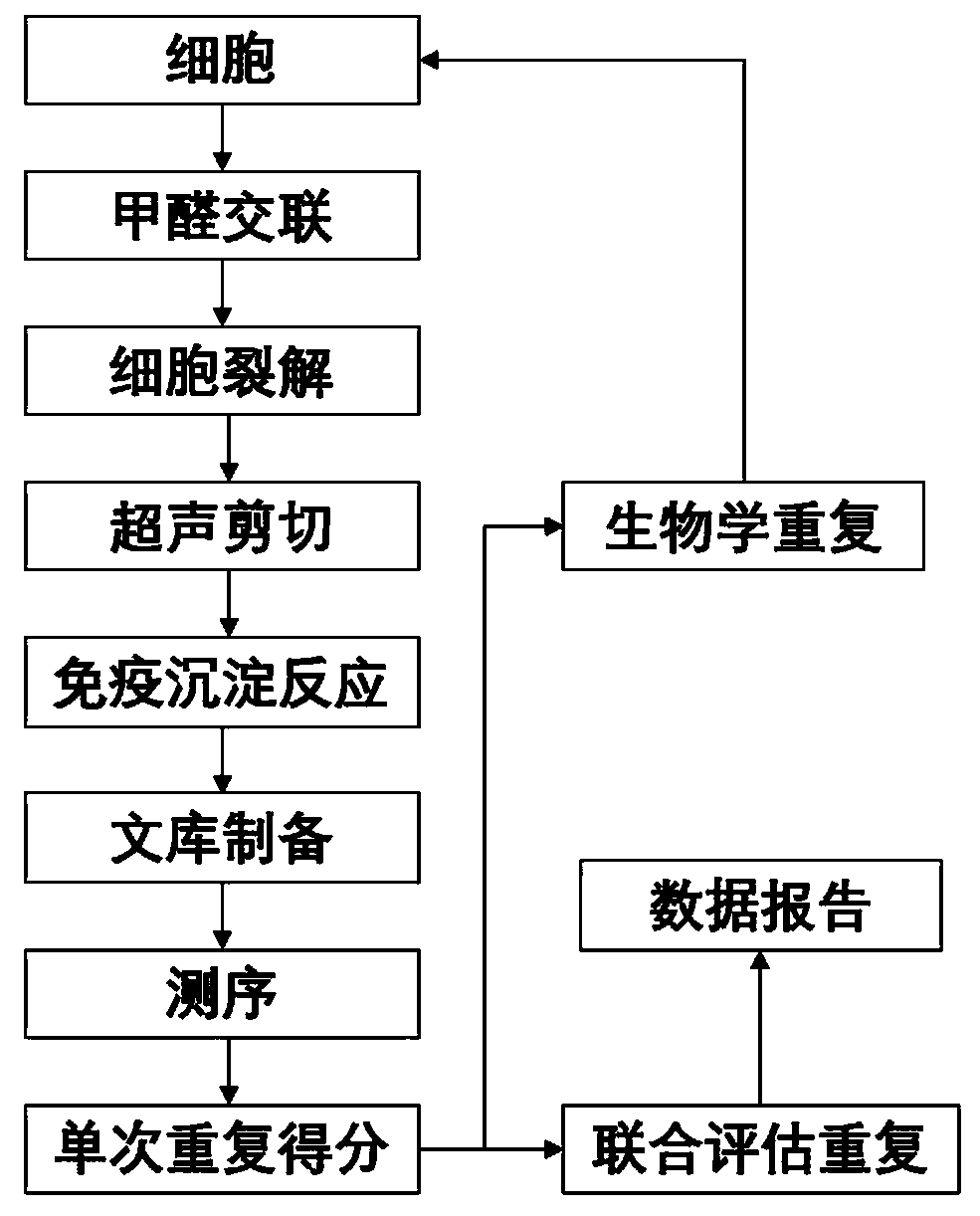
Patents
Literature
63 results about "Chromatin immunoprecipitation" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
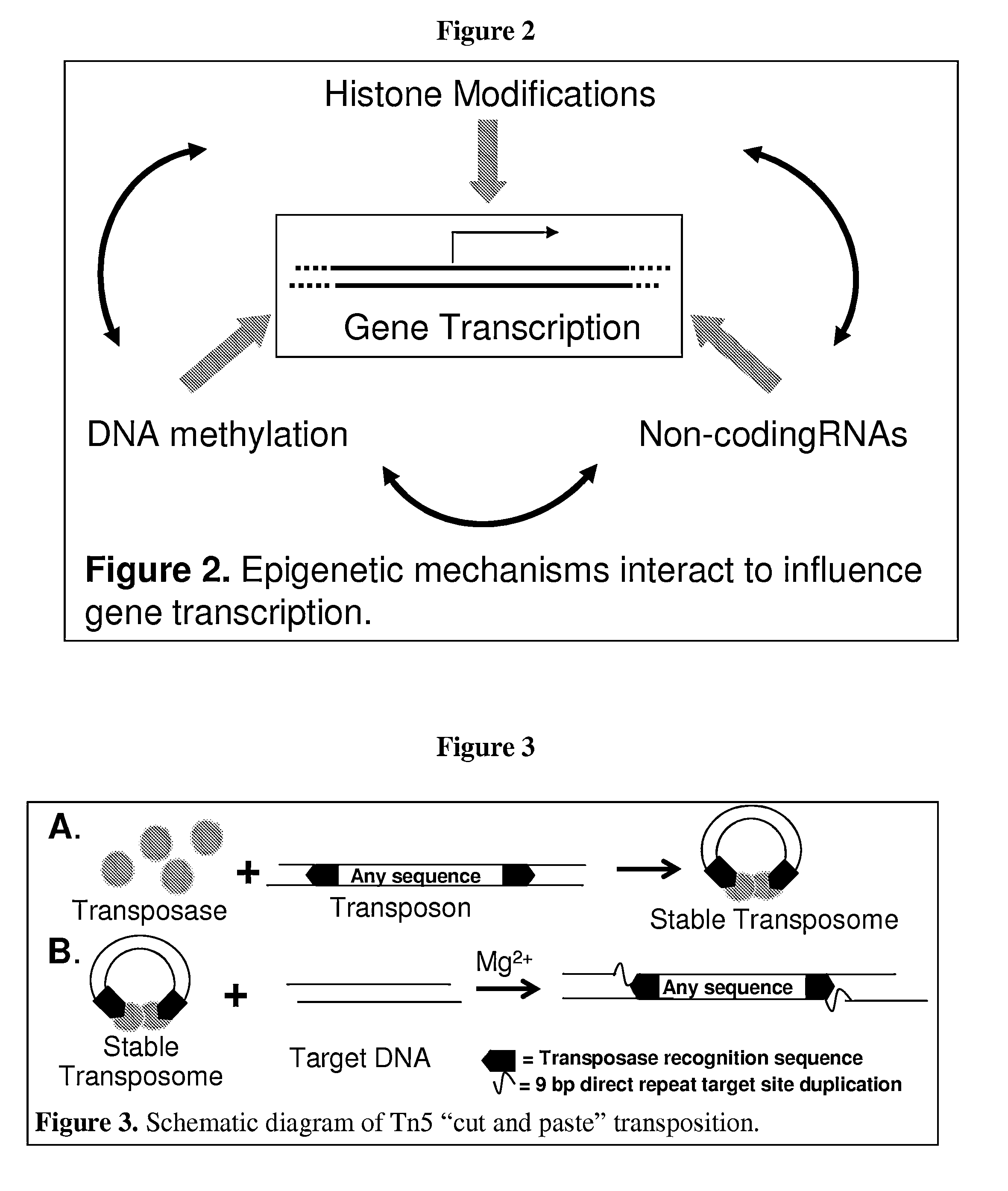
Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as transcription factors on promoters or other DNA binding sites, and possibly defining cistromes. ChIP also aims to determine the specific location in the genome that various histone modifications are associated with, indicating the target of the histone modifiers.
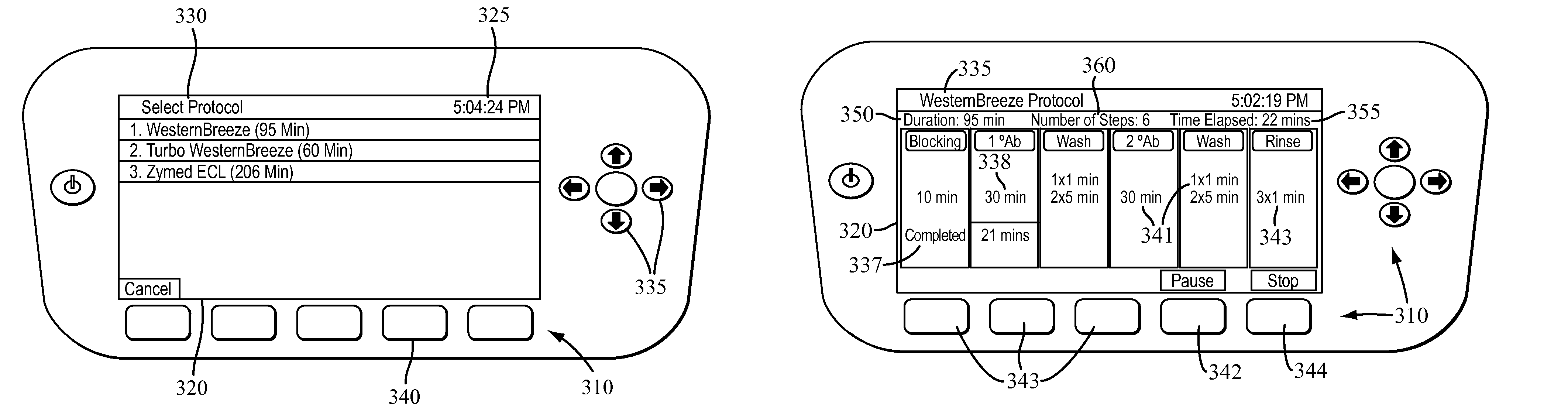
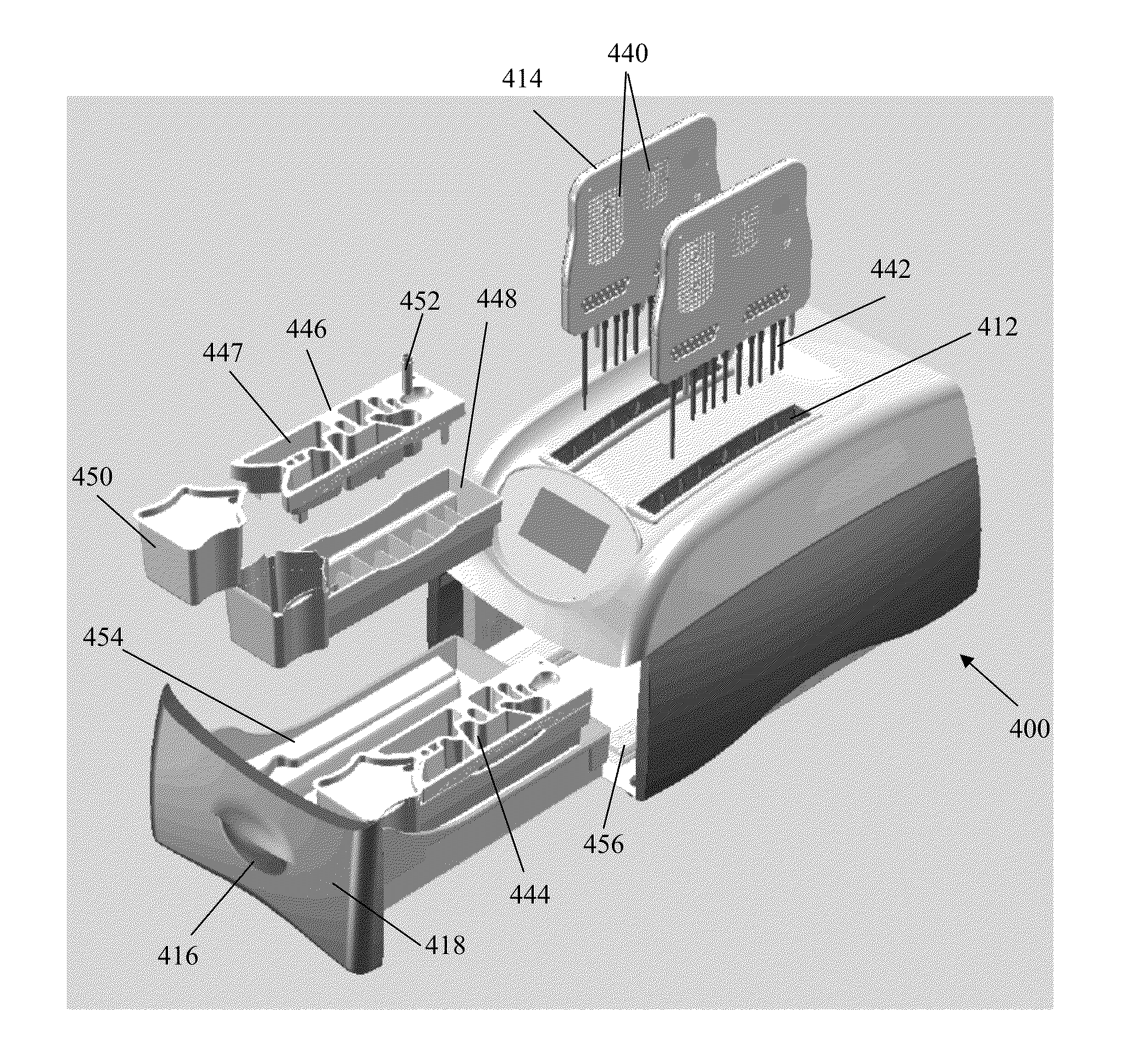
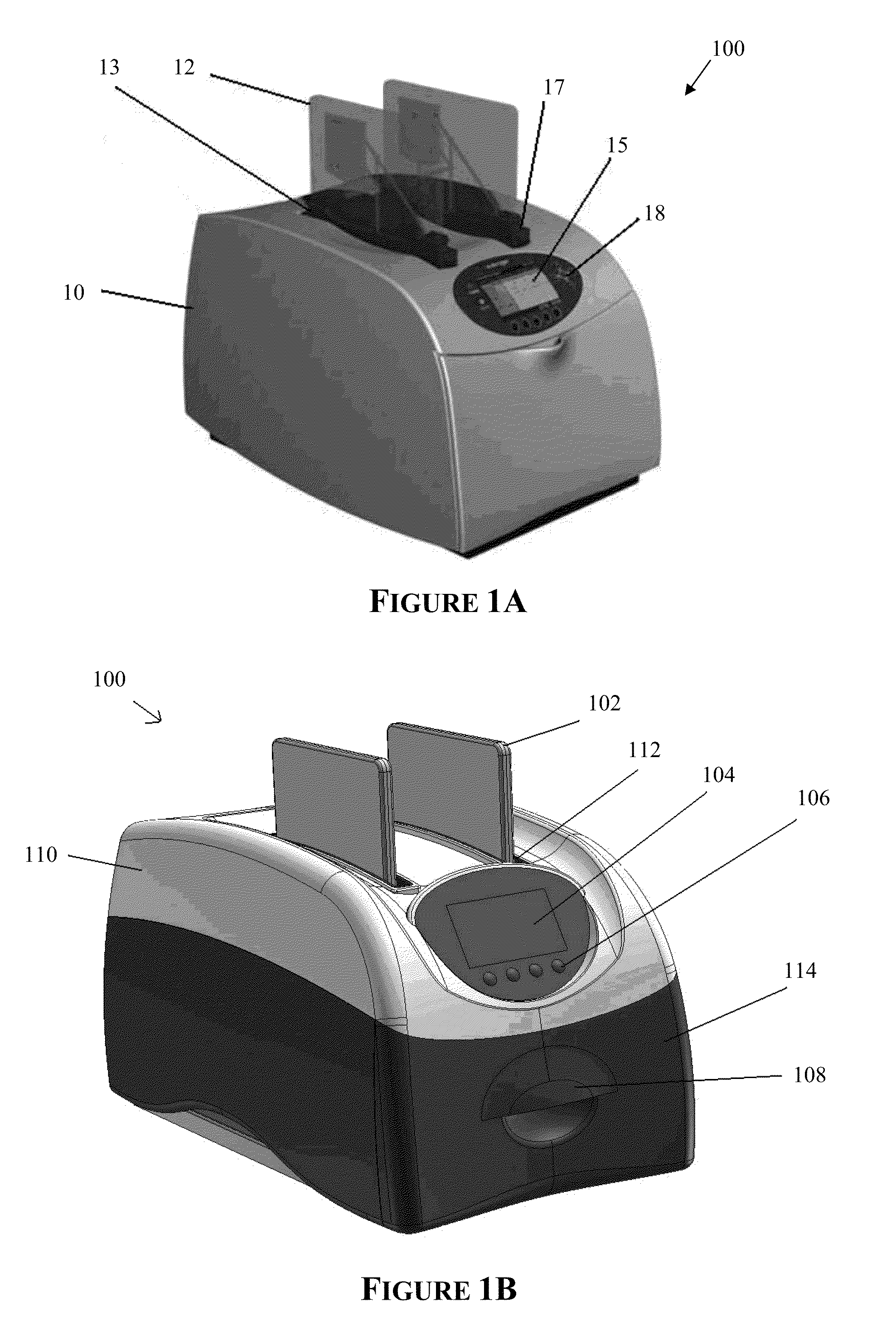
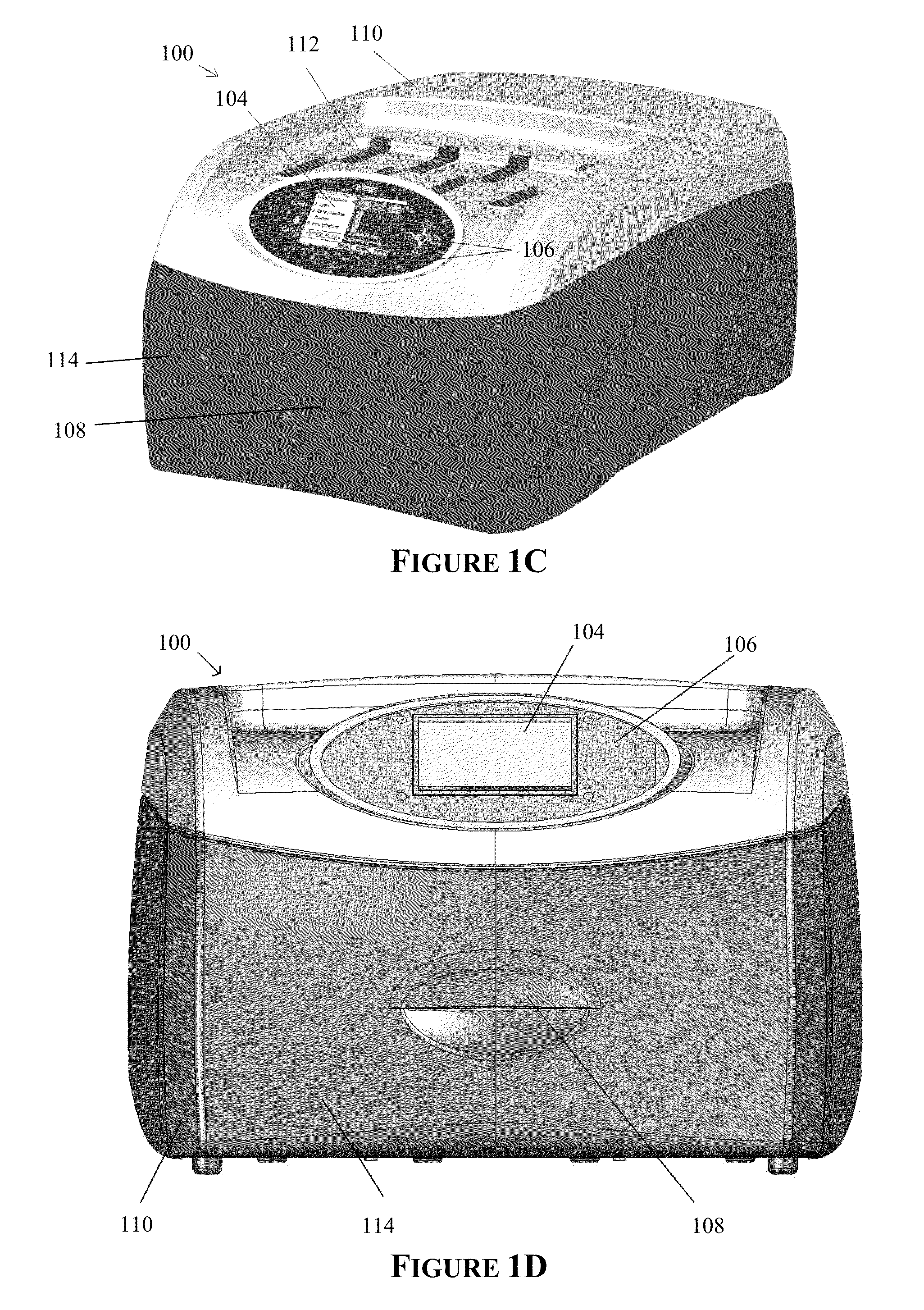
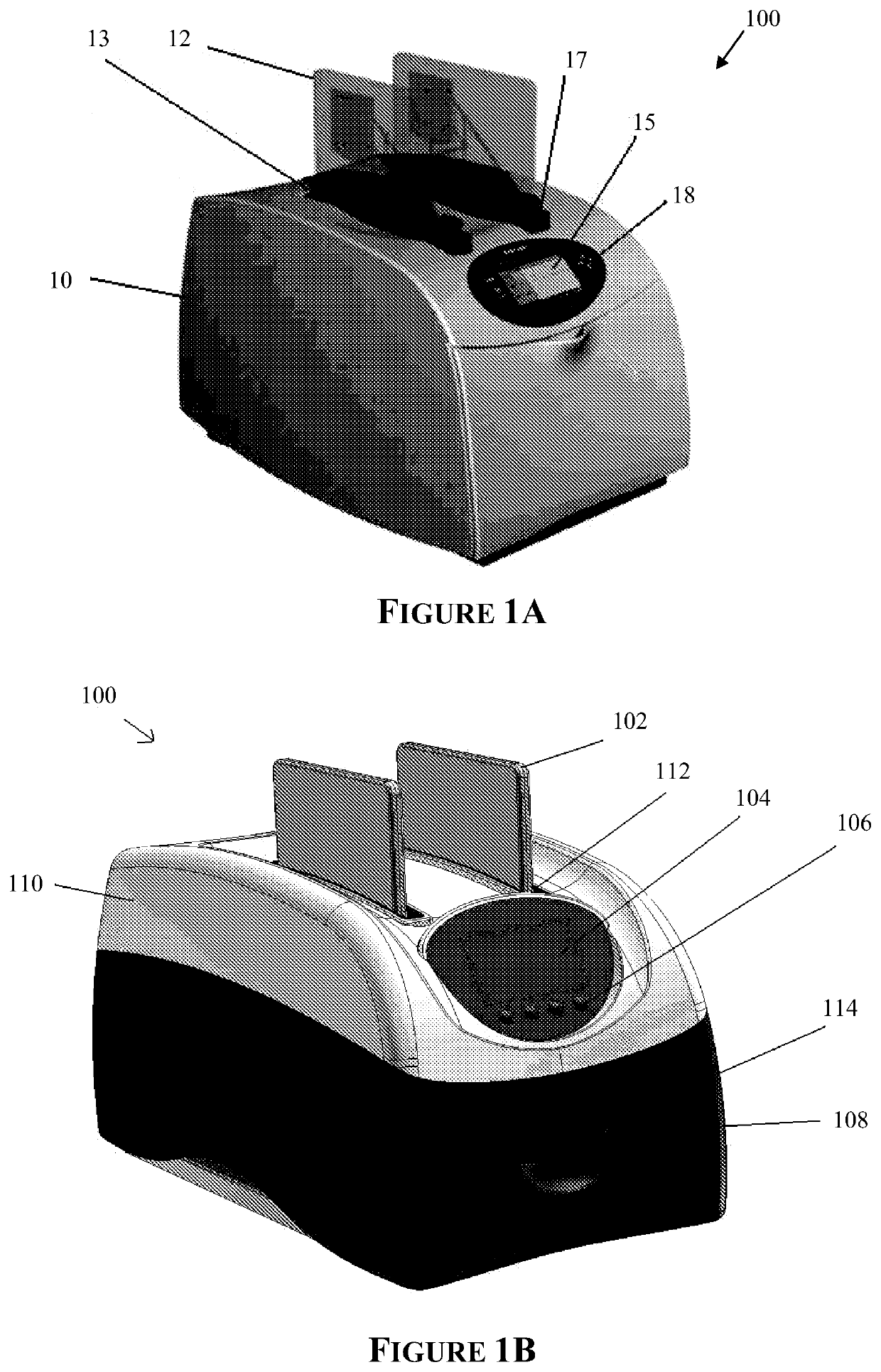
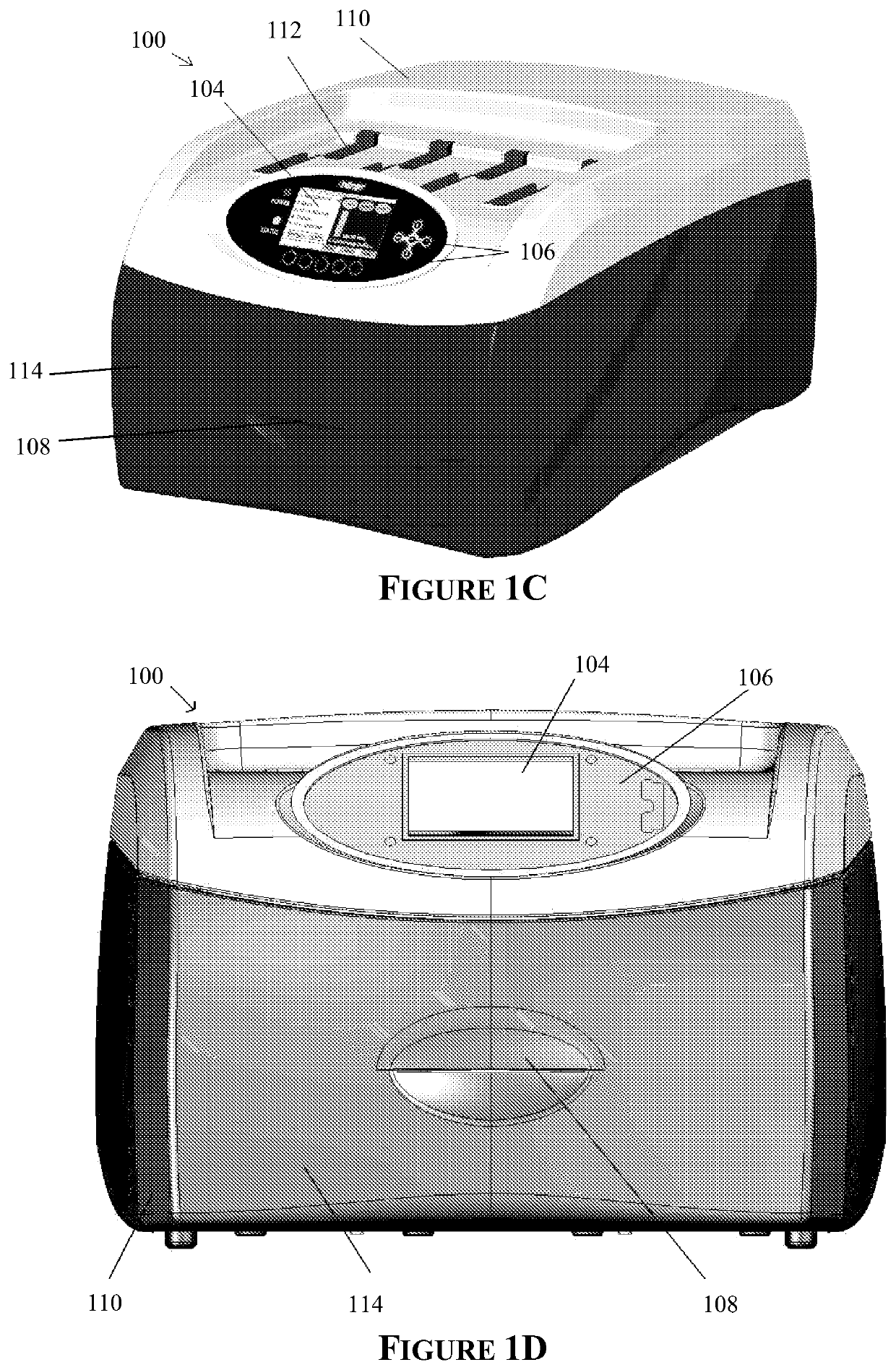
Apparatus for and method of processing biological samples
The present invention provides systems, devices, apparatuses and methods for automated bioprocessing. Examples of protocols and bioprocessing procedures suitable for the present invention include but are not limited to: immunoprecipitation, chromatin immunoprecipitation, recombinant protein isolation, nucleic acid separation and isolation, protein labeling, separation and isolation, cell separation and isolation, food safety analysis and automatic bead based separation. In some embodiments, the invention provides automated systems, automated devices, automated cartridges and automated methods of western blot processing. Other embodiments include automated systems, automated devices, automated cartridges and automated methods for separation, preparation and purification of nucleic acids, such as DNA or RNA or fragments thereof, including plasmid DNA, genomic DNA, bacterial DNA, viral DNA and any other DNA, and for automated systems, automated devices, automated cartridges and automated methods for processing, separation and purification of proteins, peptides and the like.
Owner:LIFE TECH CORP
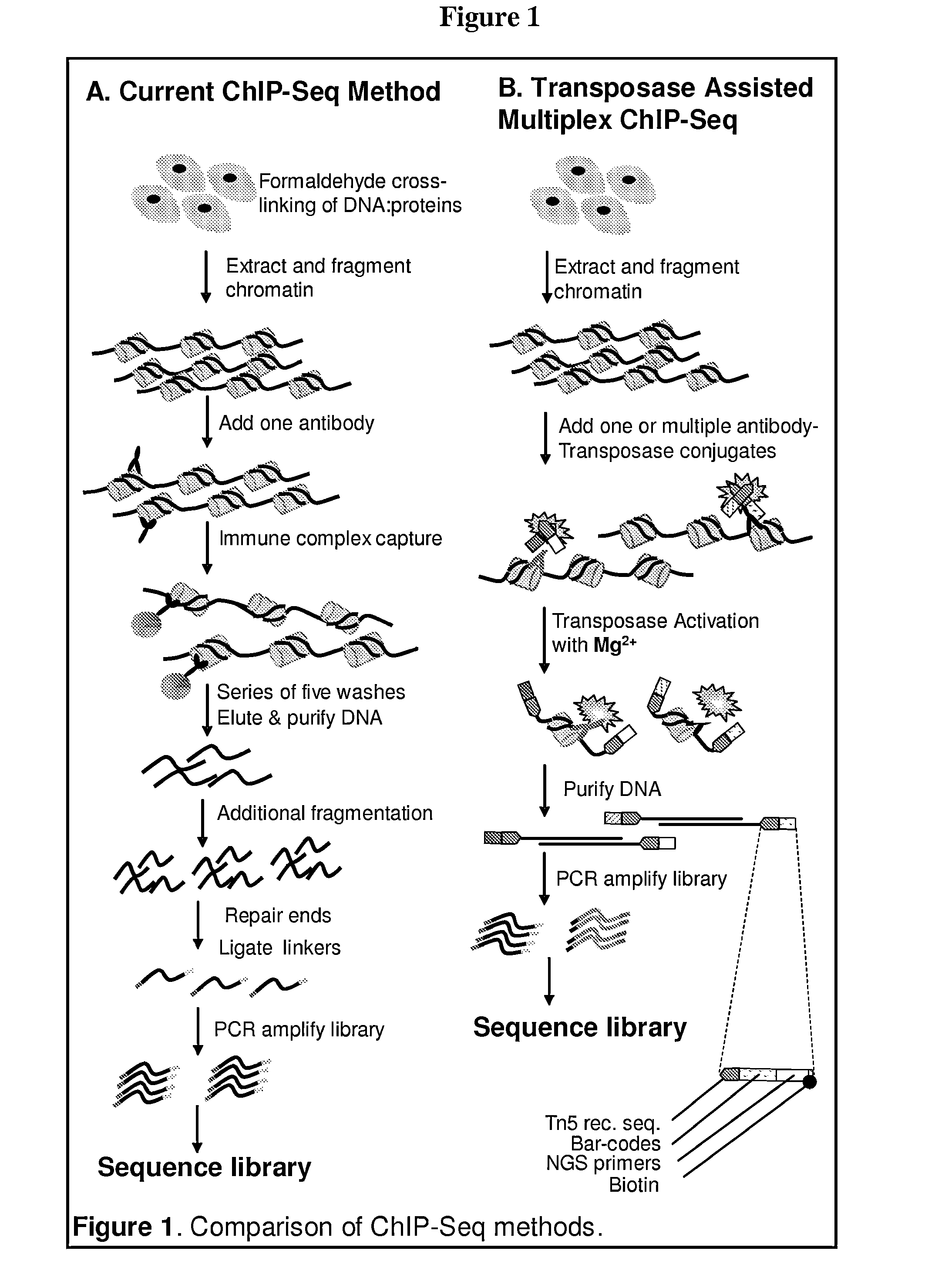
Multiplex isolation of protein-associated nucleic acids
ActiveUS20150111788A1Microbiological testing/measurementEnzyme stabilisationGenomeChromatin immunoprecipitation
The invention provides novel methods and materials for genetic and genomic analysis using single or multiplex isolation of protein-associated nucleic acids, including transposase-assisted chromatin immunoprecipitation (TAM-ChIP) and antibody-oligonucleotide proximity ligation. These methods comprise tagging and isolating chromatin or other protein-associated nucleic acids and using antibody-oligonucleotide complexes that recognize the proteins associated with such nucleic acids.
Owner:ACTIVE MOTIF INC
Method of identifying active chromatin domains
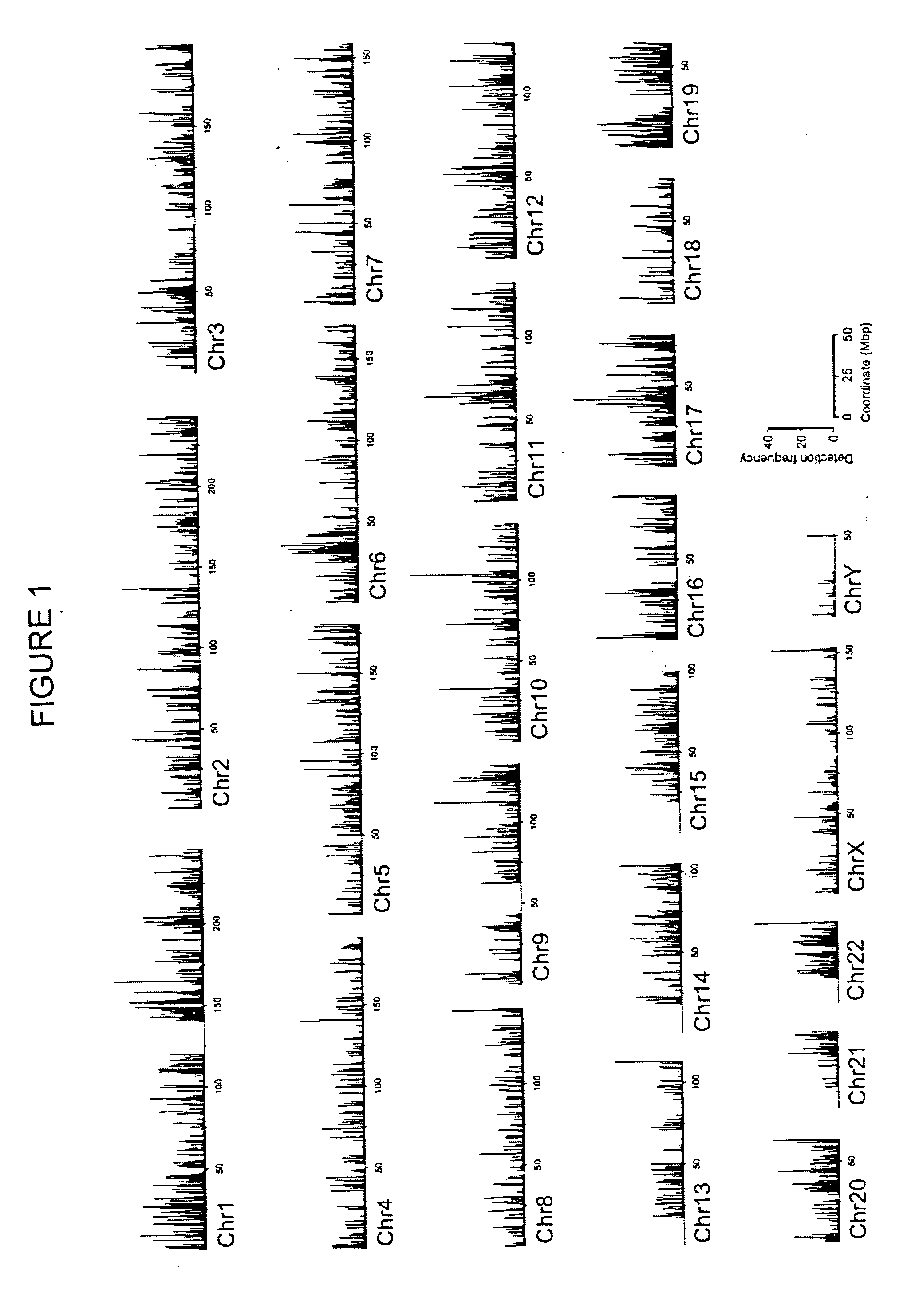
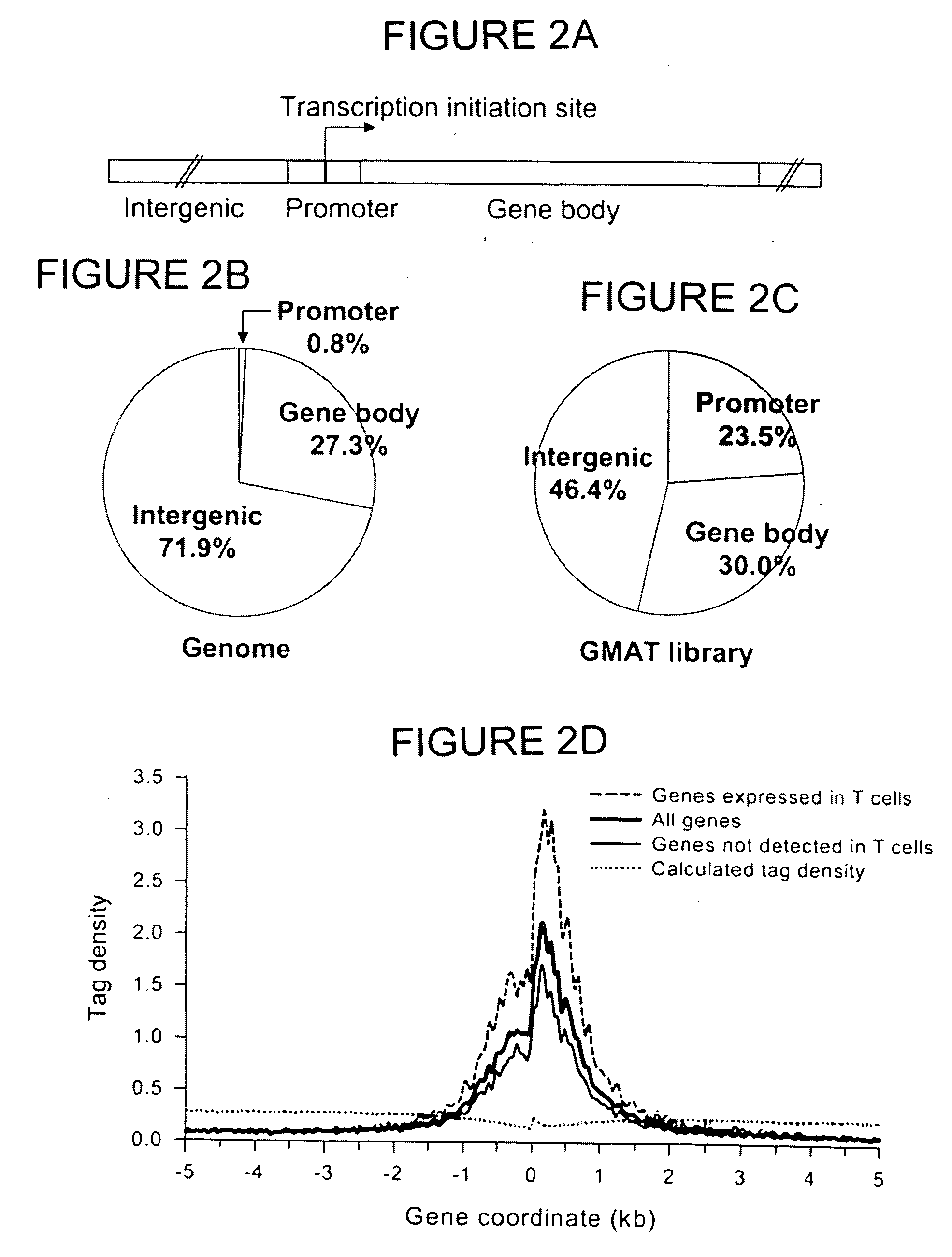
The invention provides a method of mapping DNA-protein interactions within a genome by fixing living cells to cross-link DNA and proteins, lysing the cells, and isolating chromatin by immunoprecipitation. DNA is purified and a SAGE protocol is performed on the purified DNA to produce GMAT-tag sequences, which are compared to a genomic sequence of the living cells to map DNA-protein interactions. The invention further provides a method of identifying an active chromatin domain and a method of identifying aberrant chromatin acetylation, wherein chromatin immunoprecipitation is performed using an antibody recognizing acetylated histone protein.
Owner:GOVERNMENT OF THE US REPRESENTED BY THE SEC
Methods For Preparing Sequencing Libraries
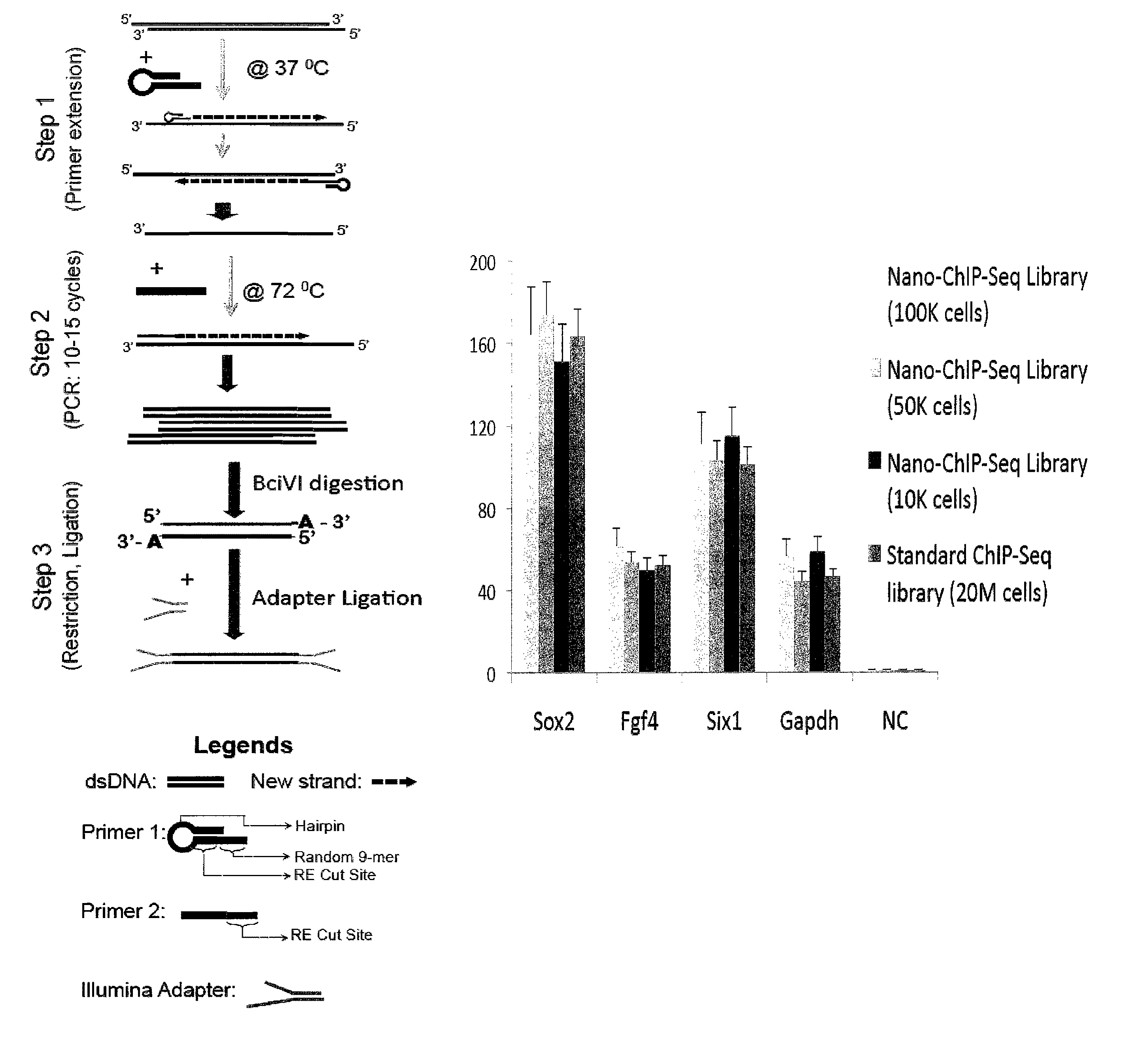
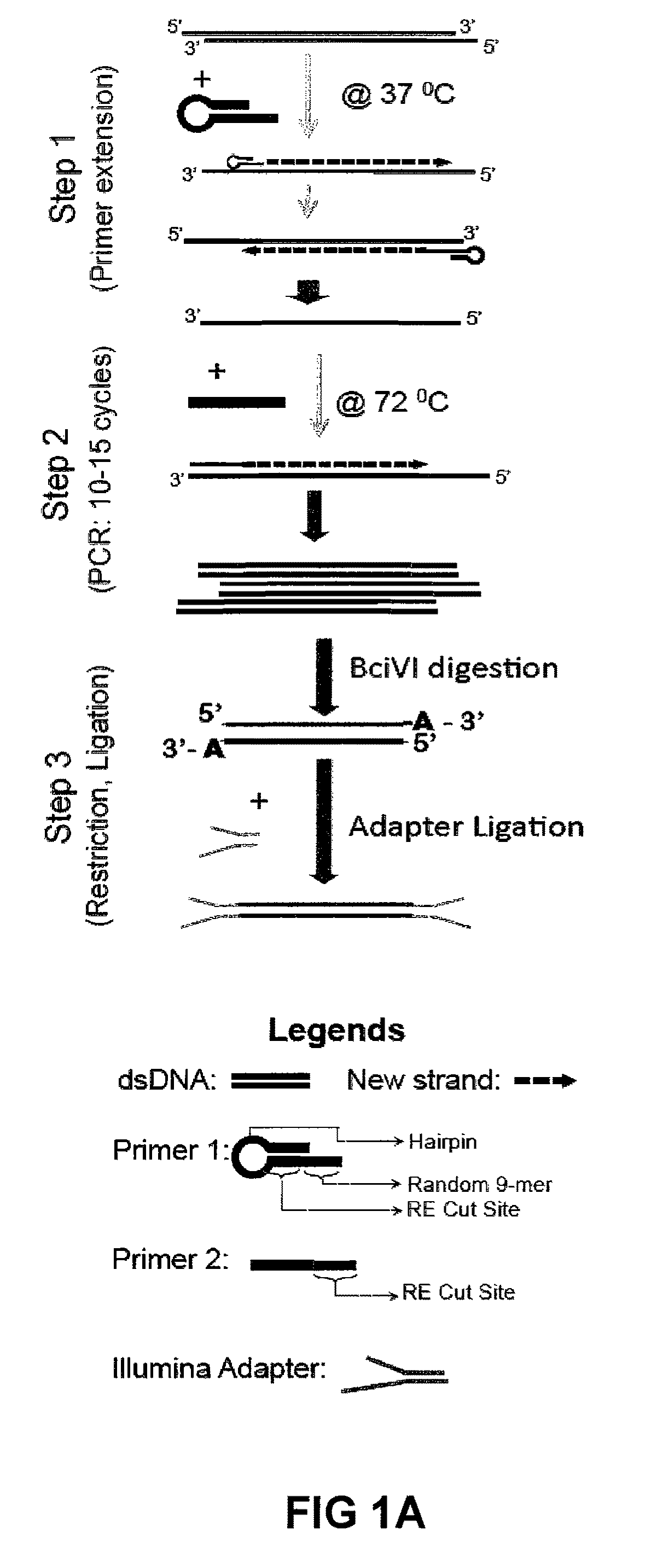
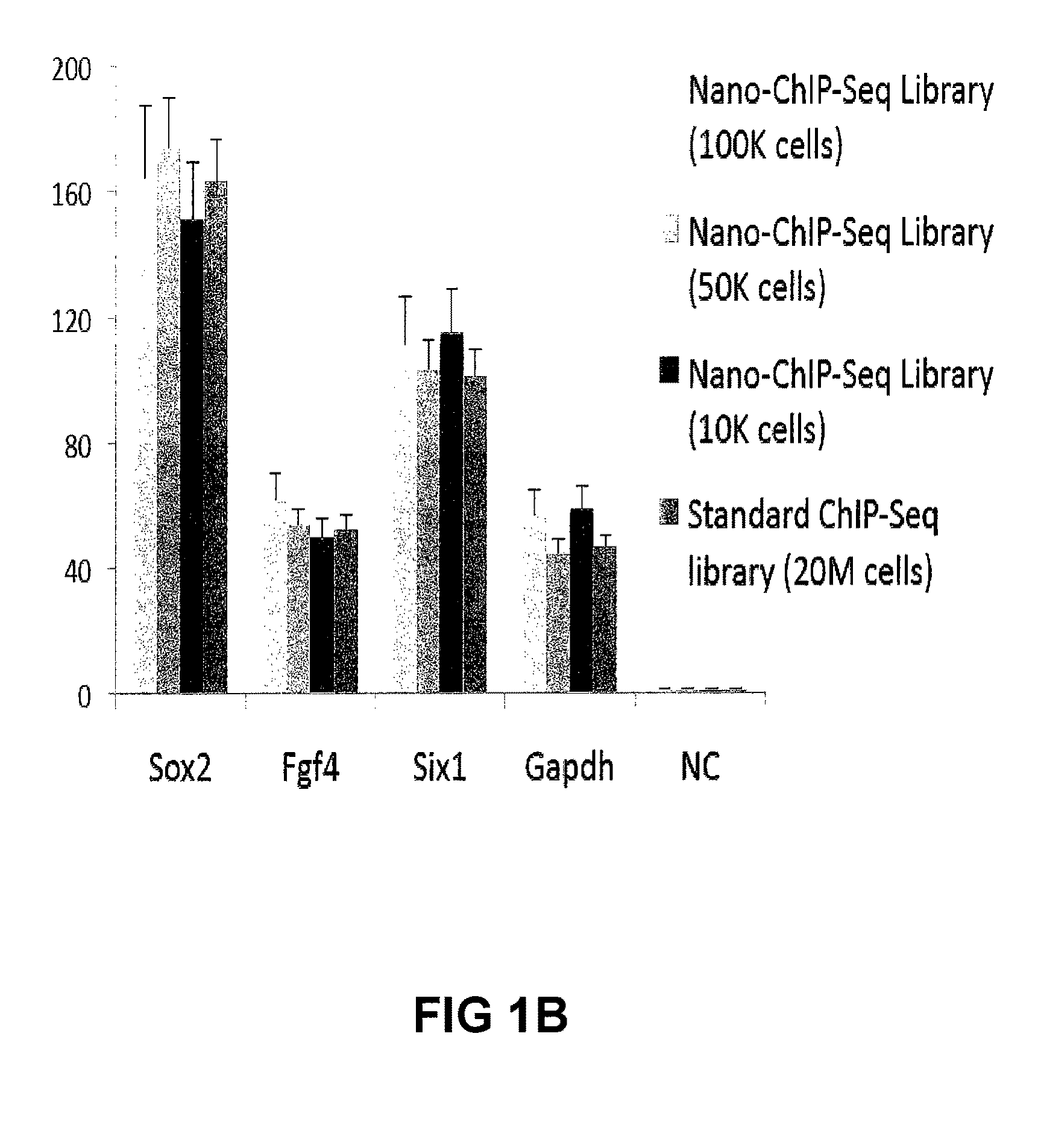
ActiveUS20110189677A1Reduce the amount requiredSugar derivativesMicrobiological testing/measurementImproved methodOrder of magnitude
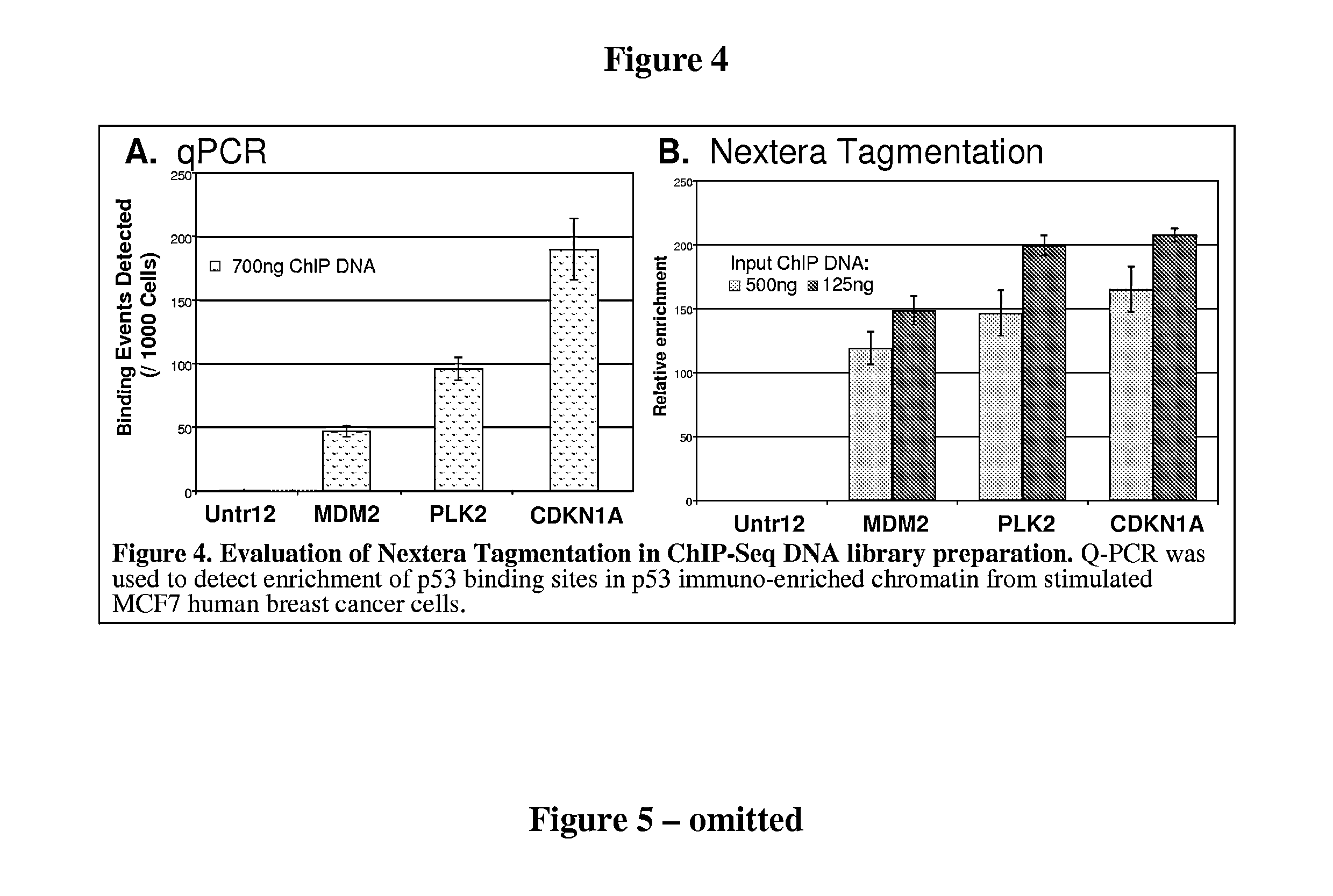
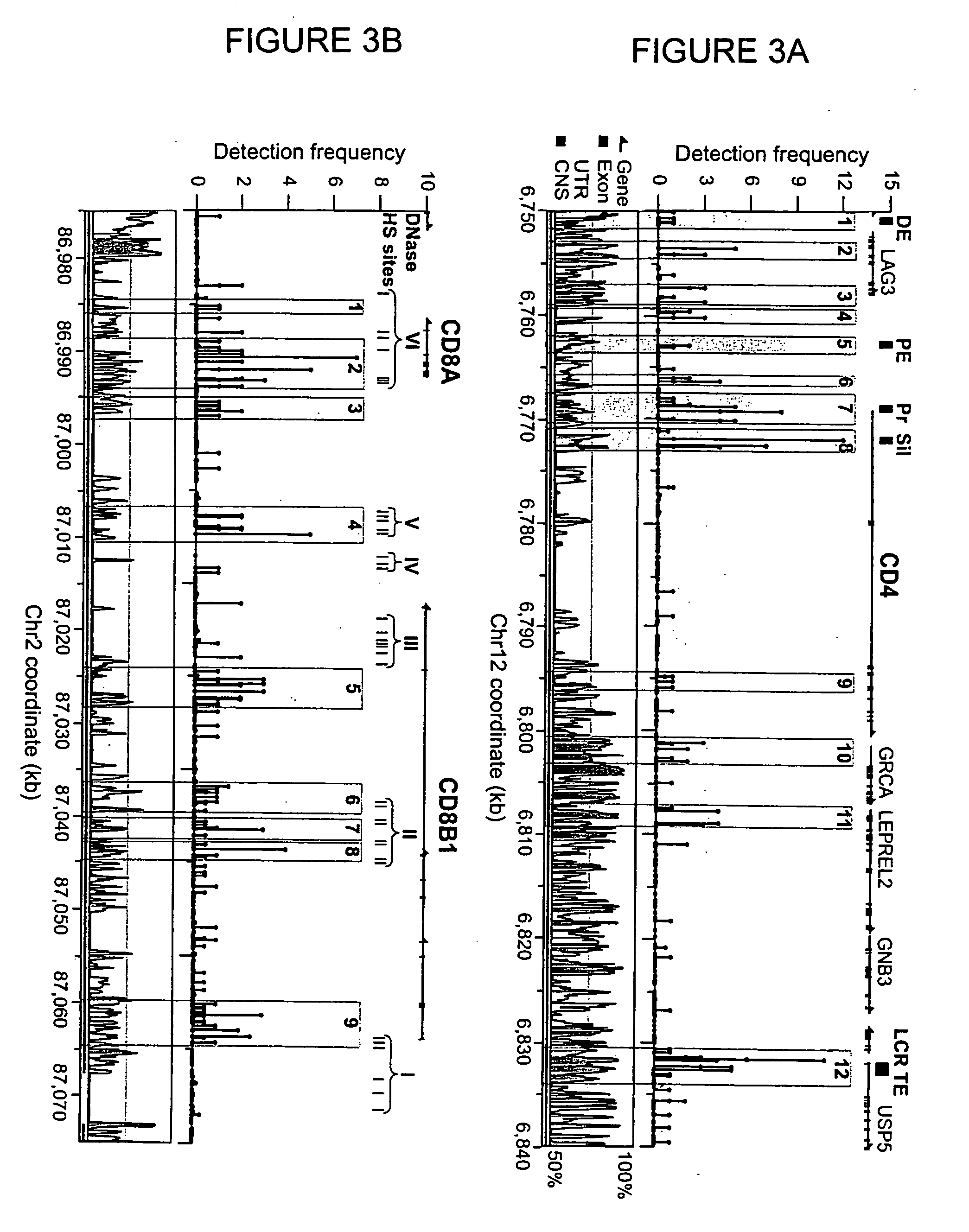
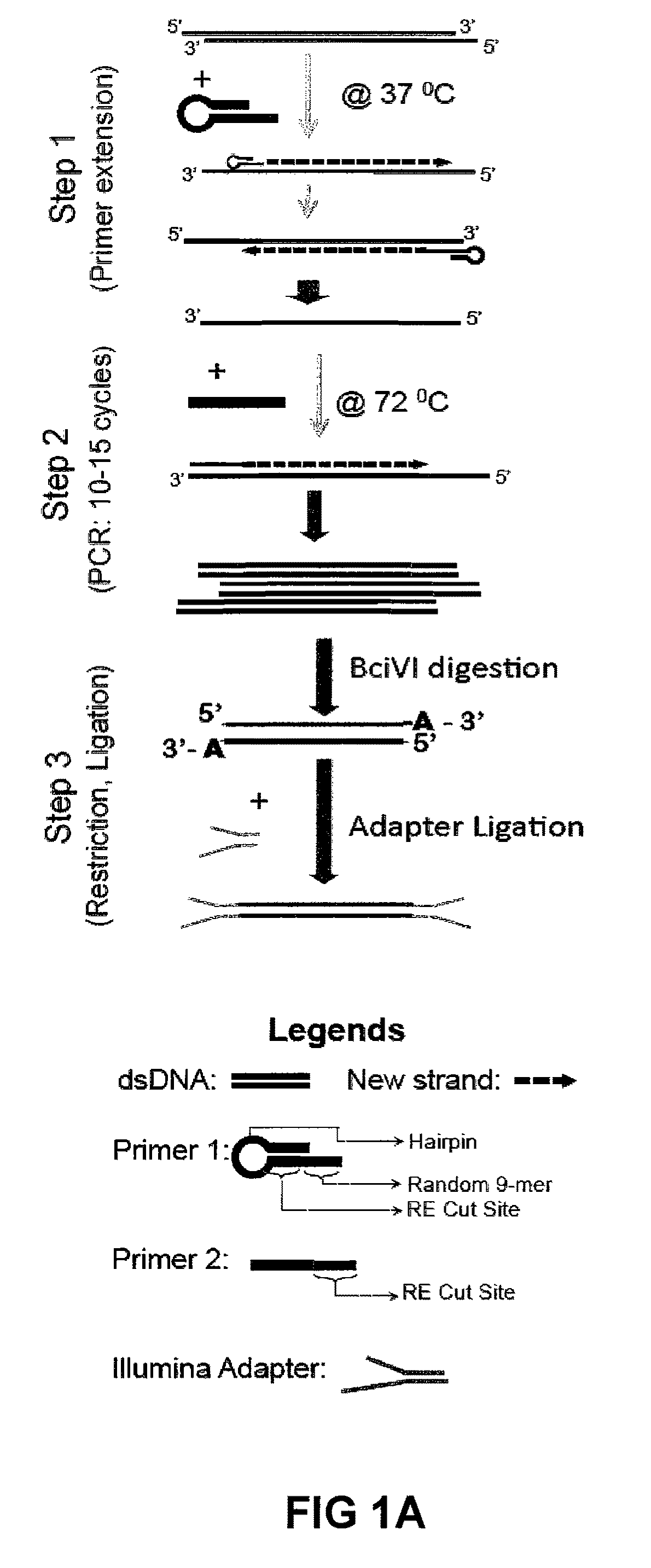
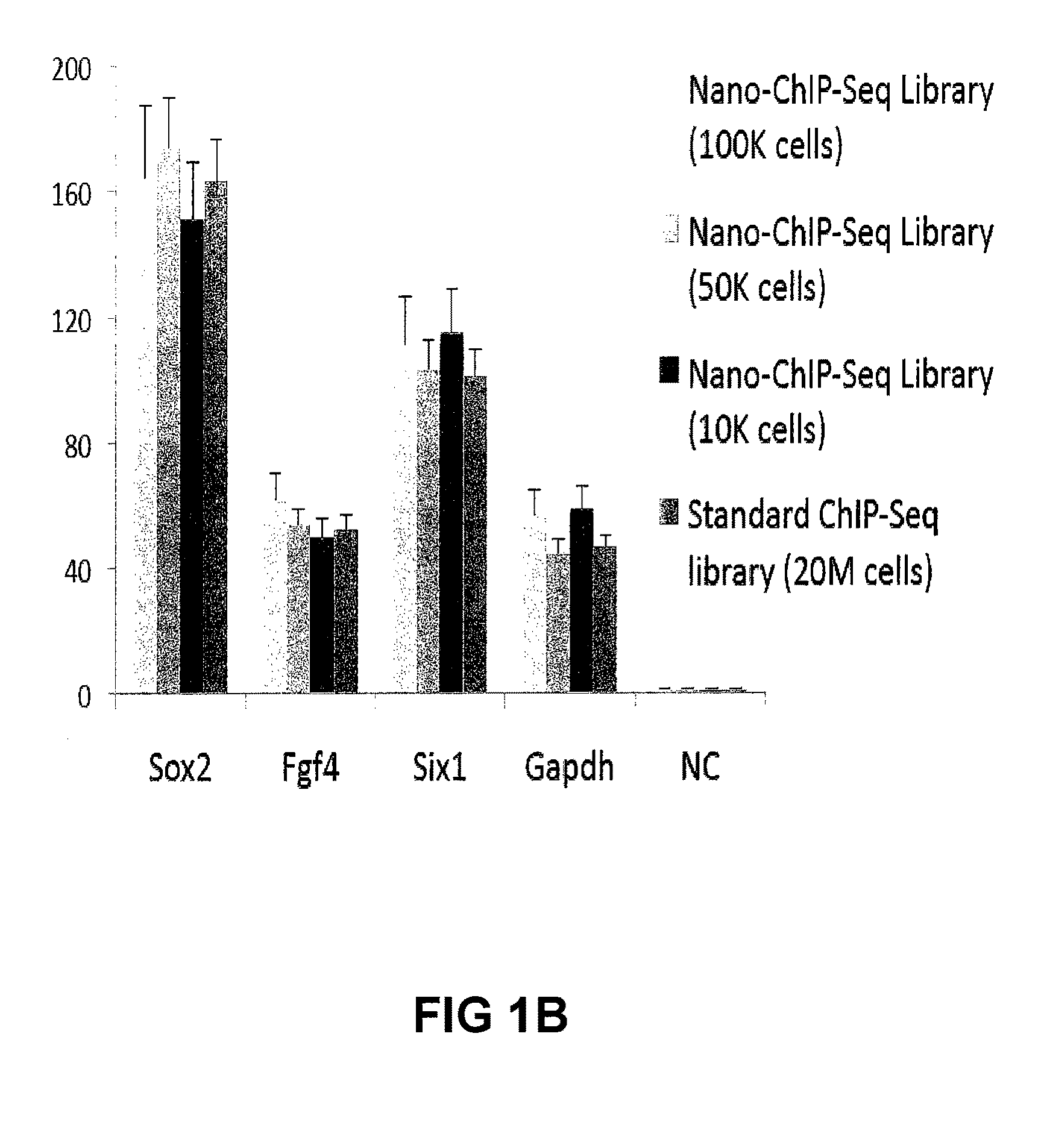
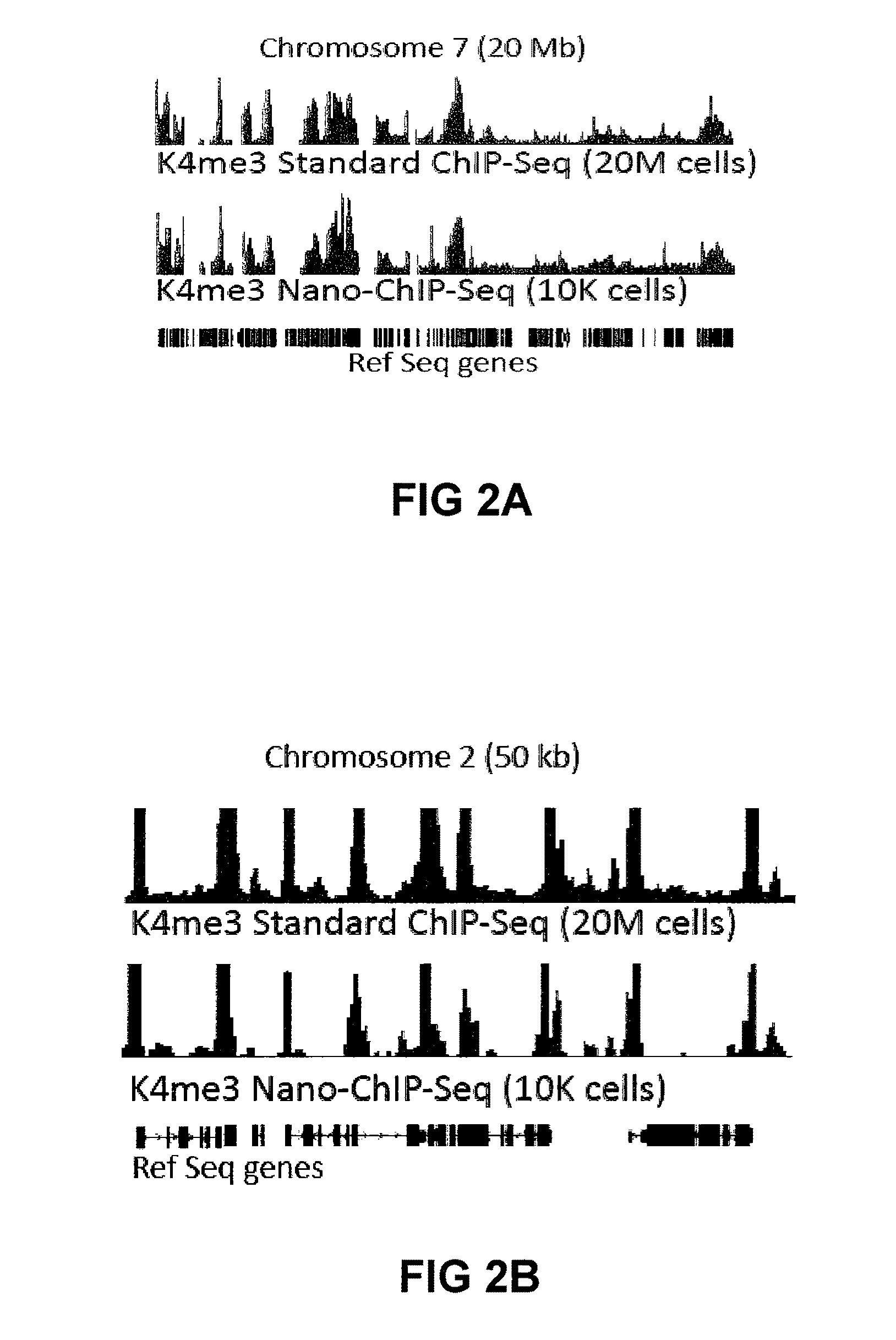
Improvements in chromatin immunoprecipitation-high throughput sequencing techniques has allowed the creation of chromatin maps from limited biological sample sizes that cannot be evaluated using conventional chromatin immunoprecipitation-sequencing protocols. For example, a modified universal primer is utilized that incorporates restriction enzymes into chromatin immunoprecipitation fragments before amplification. The improved method allows the sample sizes to be several orders of magnitude less than that required for standard ChIP-Seq techniques.
Owner:THE GENERAL HOSPITAL CORP +1
Methods of rapid chromatin immunoprecipitation
InactiveUS20070141583A1Reduce degradationAchieved rapidly and efficientlyMicrobiological testing/measurementNucleic acid reductionIn vivoImmunoprecipitation
Owner:LI WEIWEI +1
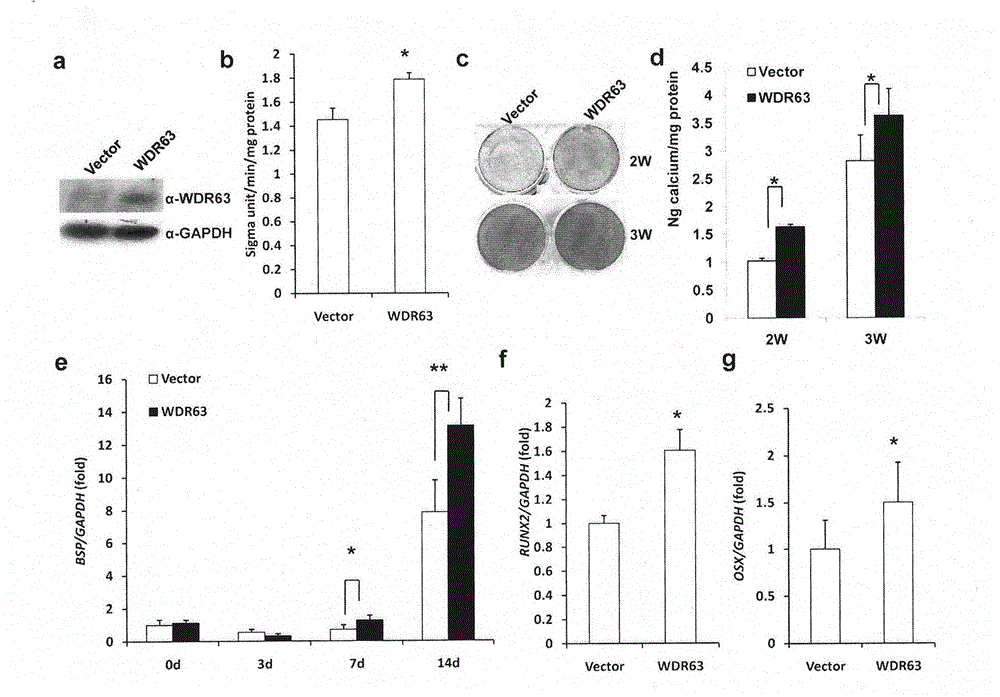
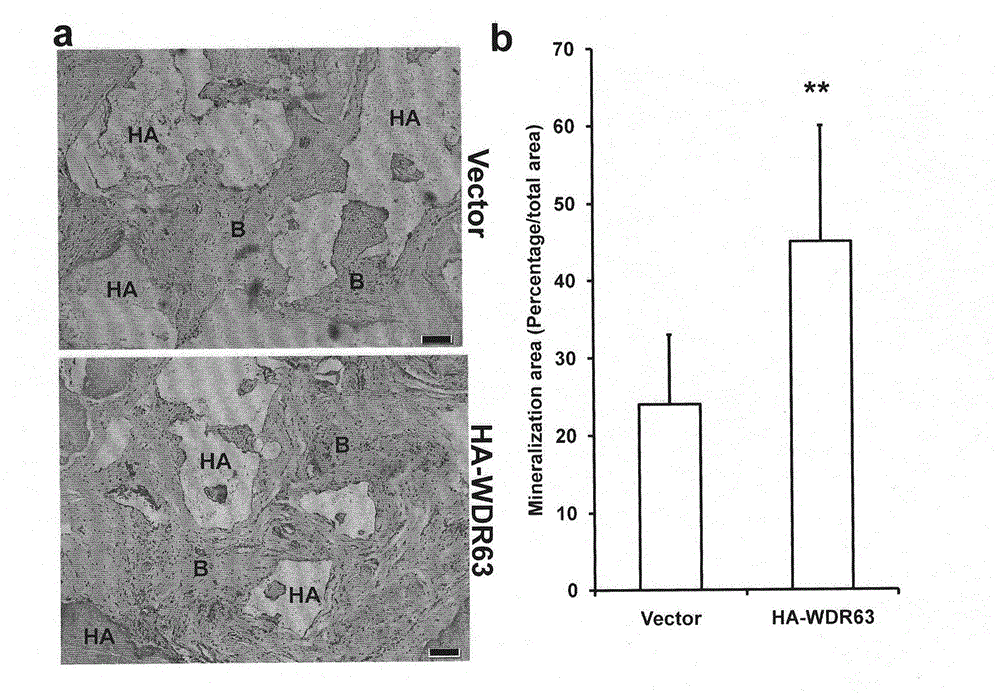
Regulating method of WDR63 gene in osteogenic differentiation and odontogenic differentiation processes of mesenchymal stem cell
ActiveCN104450621ASolve the problem of regulationFermentationGenetic engineeringReproductive capacityNude mouse
The invention discloses a regulating method of a WDR63 gene in osteogenic differentiation and odontogenicdifferentiation processes of a mesenchymal stem cell. The cell is replanted by virtue of cell culture, chromatin immunoprecipitation reaction and data analysis, plasmid construction and virus infection, western blotting analysis, cell reproductive capacity determination and alkaline phosphatase and alizarin red dyeing subcutaneously of a nude mouse, and the regulating effect of the WDR63 gene in osteogenic differentiation and odontogenic differentiation processes of the mesenchymal stem cell and the effect of promoting regeneration of tooth tissues are found. By virtue of the adjusting effect of gene activity and function of the mesenchymal stem cell by trimethylation of lysine on the fourth site of histone 3, the regulating effect of the WDR63 gene in osteogenic differentiation and odontogenicdifferentiation processes of the mesenchymal stem cell and the effect of the WDR63 gene in promoting regeneration of tooth tissues, the WDR63 gene obtained probably has a promoting effect on osteogenic differentiation of stem cells from apical papilla.
Owner:BEIJING STOMATOLOGY HOSPITAL CAPITAL MEDICAL UNIV
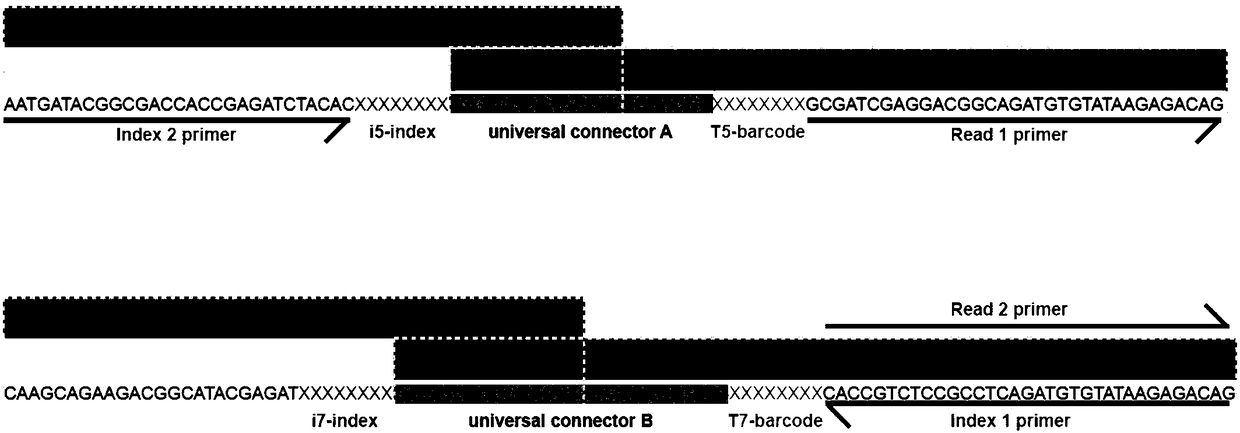
Index-first chromatin immunoprecipitation (iChIP) high-throughput sequencing experimental method applied to zebrafish embryos
InactiveCN105463090AReduce experimental biasMicrobiological testing/measurementDNA preparationAntigenEnzyme digestion
The invention relates to an index-first chromatin immunoprecipitation (iChIP) high-throughput sequencing (iChIP-seq) experimental method applied to zebrafish embryos. The method comprises the following steps: firstly, performing formaldehyde crosslinking on zebrafish embryo samples; secondly, acquiring mononucleosome through MNase (Micrococcal Nuclease) enzyme digestion; fixing chromatin on magnetic beads through H3 antigens; labeling various different samples on the magnetic beads with index joints; releasing the chromatin from the magnetic beads, concentrating, mixing the samples together, performing a chromatin immunoprecipitation reaction on various antigens to obtain DNA (Deoxyribose Nucleic Acid) sample, and purifying the DNA sample to perform library establishment and sequencing. The iChIP is an improvement on conventional ChIP (Chromatin Immunoprecipitation). According to the method, each sample is marked before ChIP, and then all the samples are mixed together to perform an experiment, so that a dynamical change decorated by the chromatin in a developing process can be observed, experimental deviations possibly caused by a small quantity of early-period cells are reduced, and the chromatin-modified dynamical change in the whole developing process can be quantitatively observed through the quantity of cells.
Owner:TONGJI UNIV
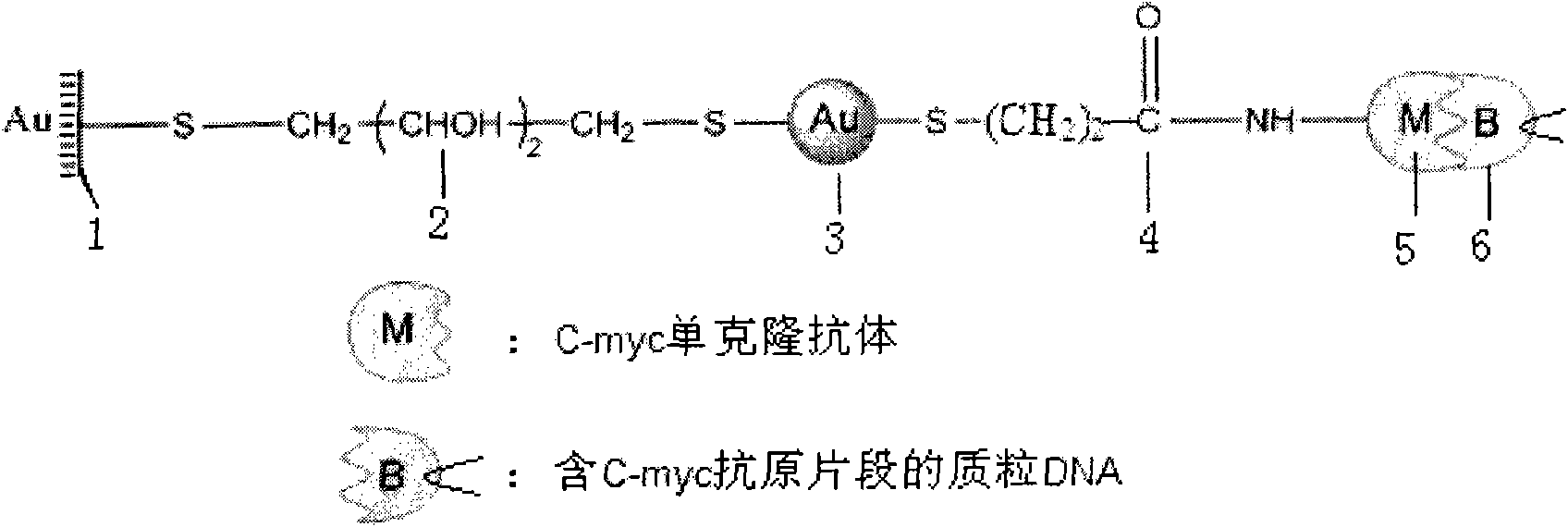
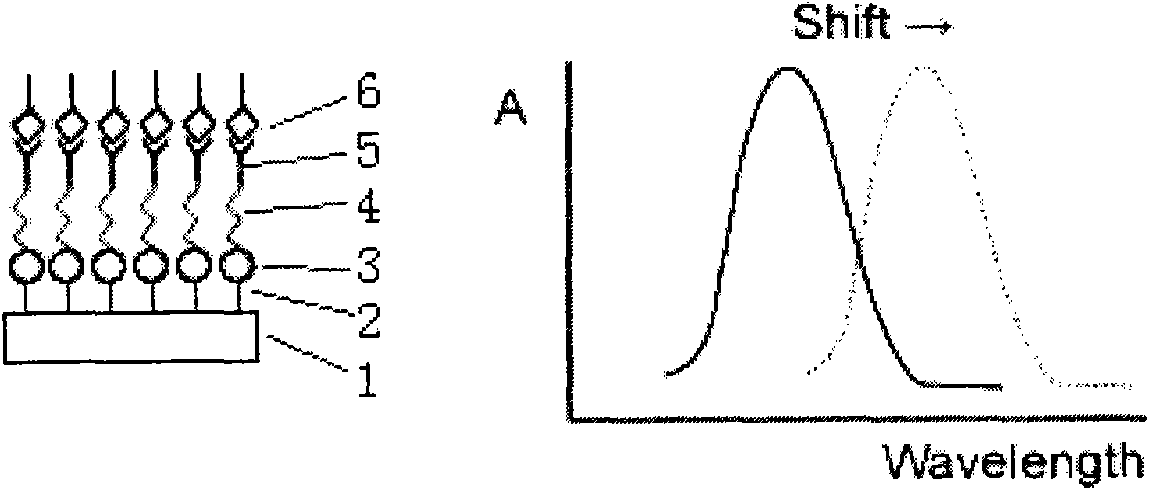
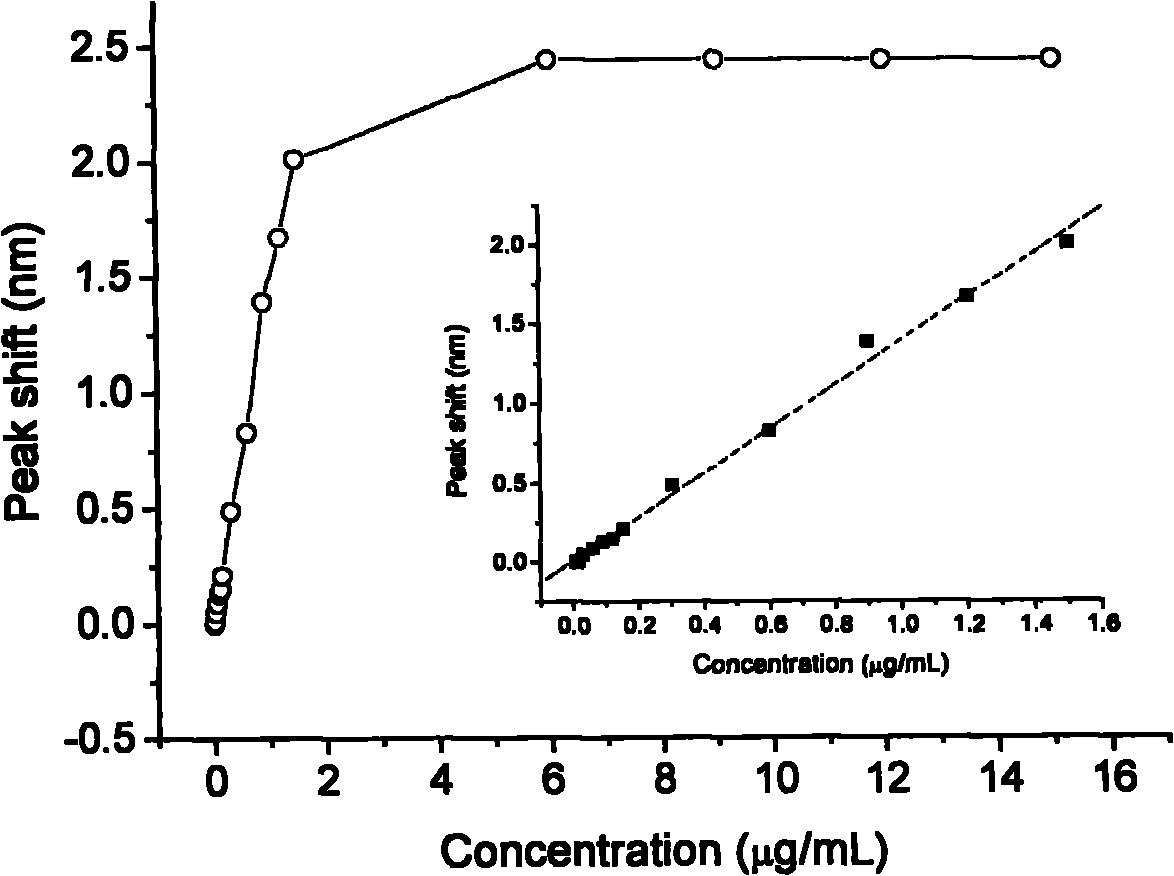
LSPR (Localized Surface Plasmon Resonance) sensing chip for detecting plasmid DNA (deoxyribonucleic acid) containing oncogene C-myc antigen fragment
The invention provides an LSPR (Localized Surface Plasmon Resonance) sensing chip for detecting plasmid DNA (deoxyribonucleic acid) containing an oncogene C-myc antigen fragment on the basis of LSPR. The LSPR sensing chip is characterized in that one DTT (Dithiothreitol) unimolecule layer (2) is assembled on the surface of a gold film (1) of the LSPR sensing chip; a gold nanoparticle layer (3) is accessed; the surface of the gold nanoparticle layer (3) is furnished with a 3-mercaptopropionic acid unimolecule layer (4); then DMAP-EDC [4-Dimethylamino pyridine-1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide] activation ensures that the 3-mercaptopropionic acid unimolecule layer (4) is combined with a C-myc monoclonal antibody (5); and the immune response combination of the C-myc monoclonal antibody (5) and the plasmid DNA (6) containing the C-myc antigen fragment causes that the surface of the gold nanoparticle generates the displacement of a local surface plasma resonance absorption peak so as to detect the content of the plasmid DNA (6) containing the C-myc antigen fragment in canceration tissues. The invention has the beneficial effects that the LSPR sensing chip has the advantages of multiple-path detection, high sensitivity and good selectivity and repeatability, is simple and convenient to assemble and is quick to quantify. The LSPR sensing chip is superior to the LSPR sensing chip manufactured with the traditional chromatin immunoprecipitation (ChIP) method.
Owner:CHANGSHA UNIVERSITY OF SCIENCE AND TECHNOLOGY
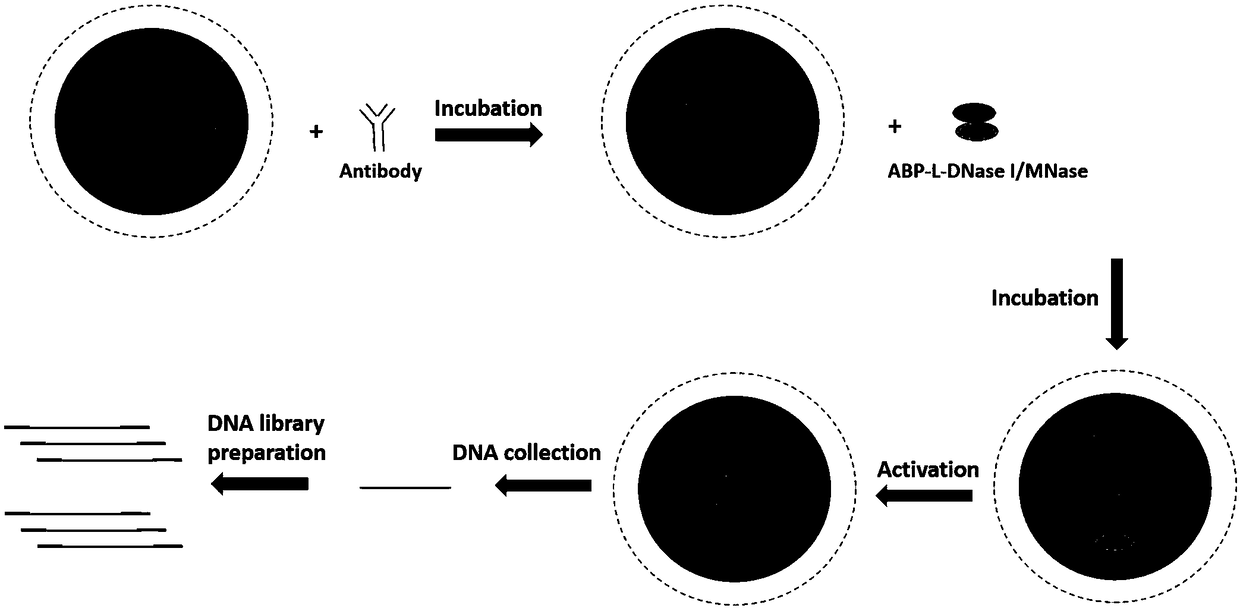
Recombinant fusion protein in antibody targeting and application thereof in epigenetics
ActiveCN109400714AReduce sequencing depthReduced affinity requirementsHydrolasesAntibody mimetics/scaffoldsEpigeneticsDNA fragmentation
The invention discloses recombinant fusion protein in antibody targeting and application thereof in epigenetics. The recombinant fusion protein comprises an ABP-L-DFE structure, wherein ABP is locatedin the N terminal of the recombinant fusion protein and represents a structure domain with a binding antibody function, L represents a connecting peptide, and DFE represents a structure domain with aDNA (Deoxyribonucleic Acid) fragmentation function. The recombinant fusion protein disclosed by the invention is capable of improving a ChIP-seq (Chromatin Immunoprecipitation-sequencing) technology,simplifying operation, increasing data quality, reducing operation difficulty and expanding the application range, particularly the application in a small amount of precious cells.
Owner:VAZYME BIOTECH NANJING
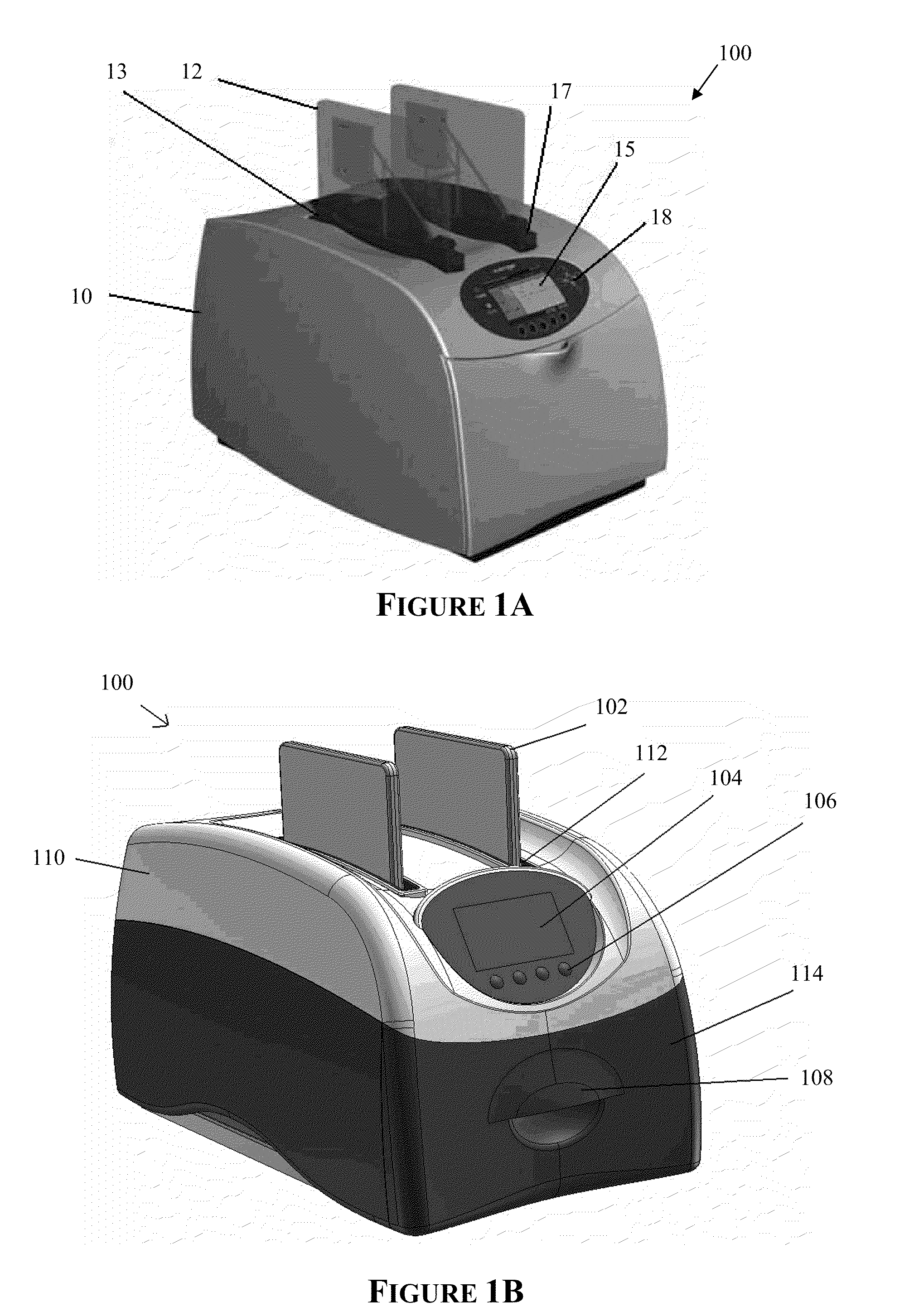
Apparatus for and method of processing biological samples
ActiveUS20130040376A1Bioreactor/fermenter combinationsHeating or cooling apparatusWestern blotGenomic DNA
The present invention provides systems, devices, apparatuses and methods for automated bioprocessing. Examples of protocols and bioprocessing procedures suitable for the present invention include but are not limited to: immunoprecipitation, chromatin immunoprecipitation, recombinant protein isolation, nucleic acid separation and isolation, protein labeling, separation and isolation, cell separation and isolation, food safety analysis and automatic bead based separation. In some embodiments, the invention provides automated systems, automated devices, automated cartridges and automated methods of western blot processing. Other embodiments include automated systems, automated devices, automated cartridges and automated methods for separation, preparation and purification of nucleic acids, such as DNA or RNA or fragments thereof, including plasmid DNA, genomic DNA, bacterial DNA, viral DNA and any other DNA, and for automated systems, automated devices, automated cartridges and automated methods for processing, separation and purification of proteins, peptides and the like.
Owner:LIFE TECH CORP
Method for rapidly preparing chromatin immunoprecipitation sample
ActiveCN106801096AShorten the timeHigh sensitivityMicrobiological testing/measurementMagnetic beadA-DNA
The invention discloses a method for rapidly preparing a chromatin immunoprecipitation sample. The method comprises the following steps: performing formaldehyde crosslinking on a cell to fix a complex of a protein and a nucleic acid, extracting the complex of the protein and the nucleic acid, and breaking a chromatin segment into small segments in an ultrasonic or enzymatic hydrolysis manner; suspending an antibody onto a solid-phase support, i.e. a magnetic bead, adding the obtained small segments onto the prepared magnetic bead, performing incubation to obtain a magnetic bead antigen antibody complex, washing an uncombined substance away after incubation is finished, and resuspending the magnetic bead antigen antibody complex to obtain a magnetic bead antigen antibody complex sample to be detected. The method is easy to operate; the prepared magnetic bead antigen antibody complex sample can be directly used for PCR detection or amplification and construction of a DNA library, so that the sample preparation time is obviously shortened, meanwhile, the shortcoming of weaker enrichment signals of certain proteins and DNAs caused by excessive elution steps is overcome, the detection sensitivity is greatly improved, and influence on subsequent PCR identification and DNA segment amplification is avoided.
Owner:WUHAN UNIV
Method of identifying active chromatin domains
The invention provides a method of mapping DNA-protein interactions within a genome by fixing living cells to cross-link DNA and proteins, lysing the cells, and isolating chromatin by immunoprecipitation. DNA is purified and a SAGE protocol is performed on the purified DNA to produce GMAT-tag sequences, which are compared to a genomic sequence of the living cells to map DNA-protein interactions. The invention further provides a method of identifying an active chromatin domain and a method of identifying aberrant chromatin acetylation, wherein chromatin immunoprecipitation is performed using an antibody recognizing acetylated histone protein.
Owner:GOVERNMENT OF THE US REPRESENTED BY THE SEC
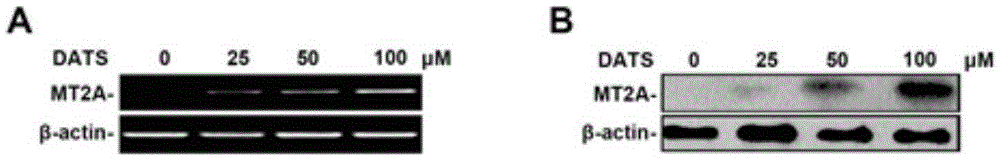
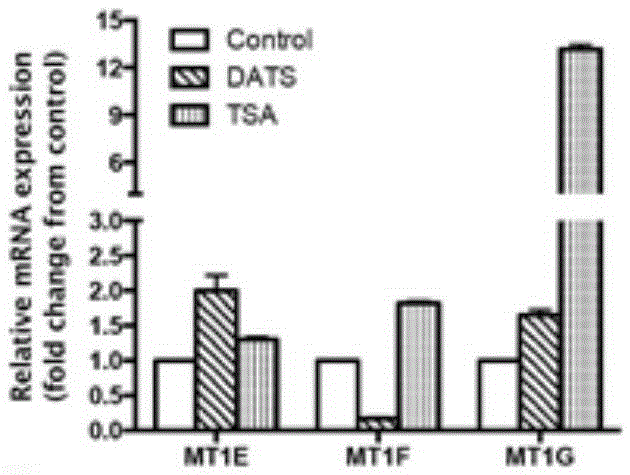
Histone acetylation modified MT2A genome detection kit and primer pairs
The invention discloses a histone acetylation modified MT2A genome detection kit and primer pairs. The detection kit comprises primer pairs as shown in SEQ ID No:2 to SEQ ID No:2 of a sequence table, a positive primer pair, a chromatin Immunoprecipitation assay kit, a nucleic acid extracting reagent and a PCR reaction agent. The histone acetylation modified MT2A genome detection kit can be used for specficially detecting histone acetylation level associated with DNA of a MT2A genome; meanwhile, the reagent kit also can be used for carrying out peculiar and quantitative detection on the genome histone acetylation level associated with the DNA of the MT2A genome; and the detected results can provide basis for establishing a scheme for treating cancer patients, and also provides an evaluation method for prognosis, on drug therapy, of the cancer patients.
Owner:BEIJING JIAOTONG UNIV
Trace cell ChIP (Chromatin Immunoprecipitation) method
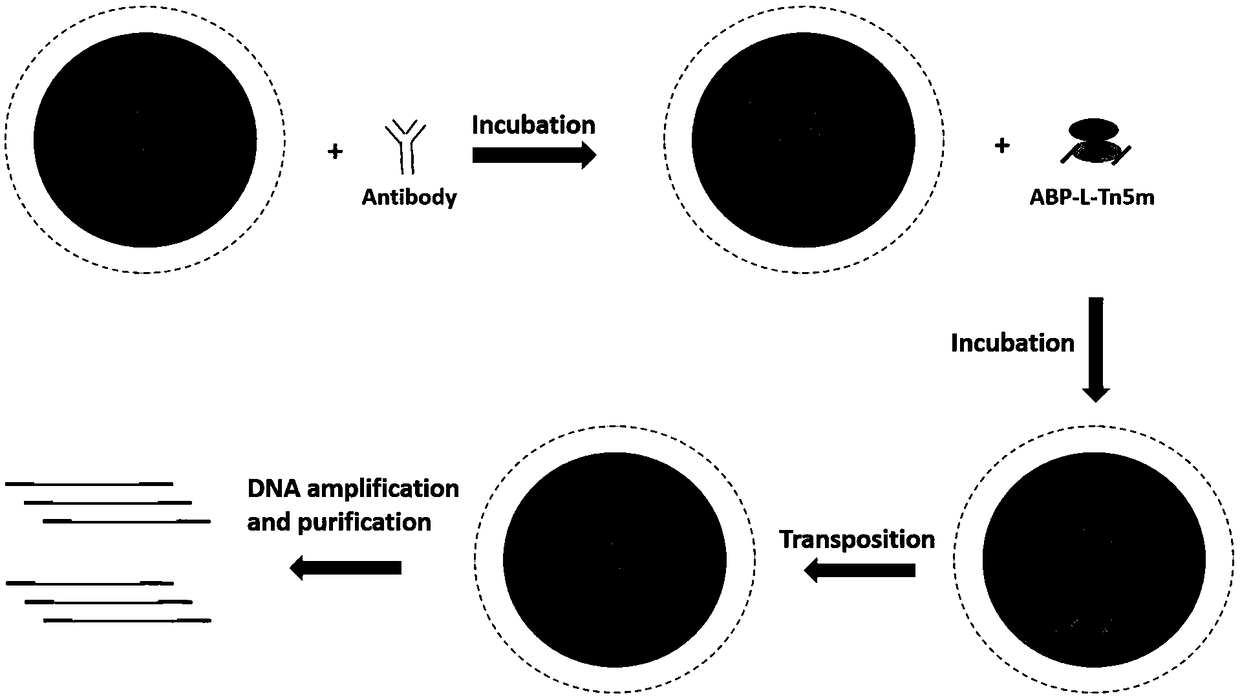
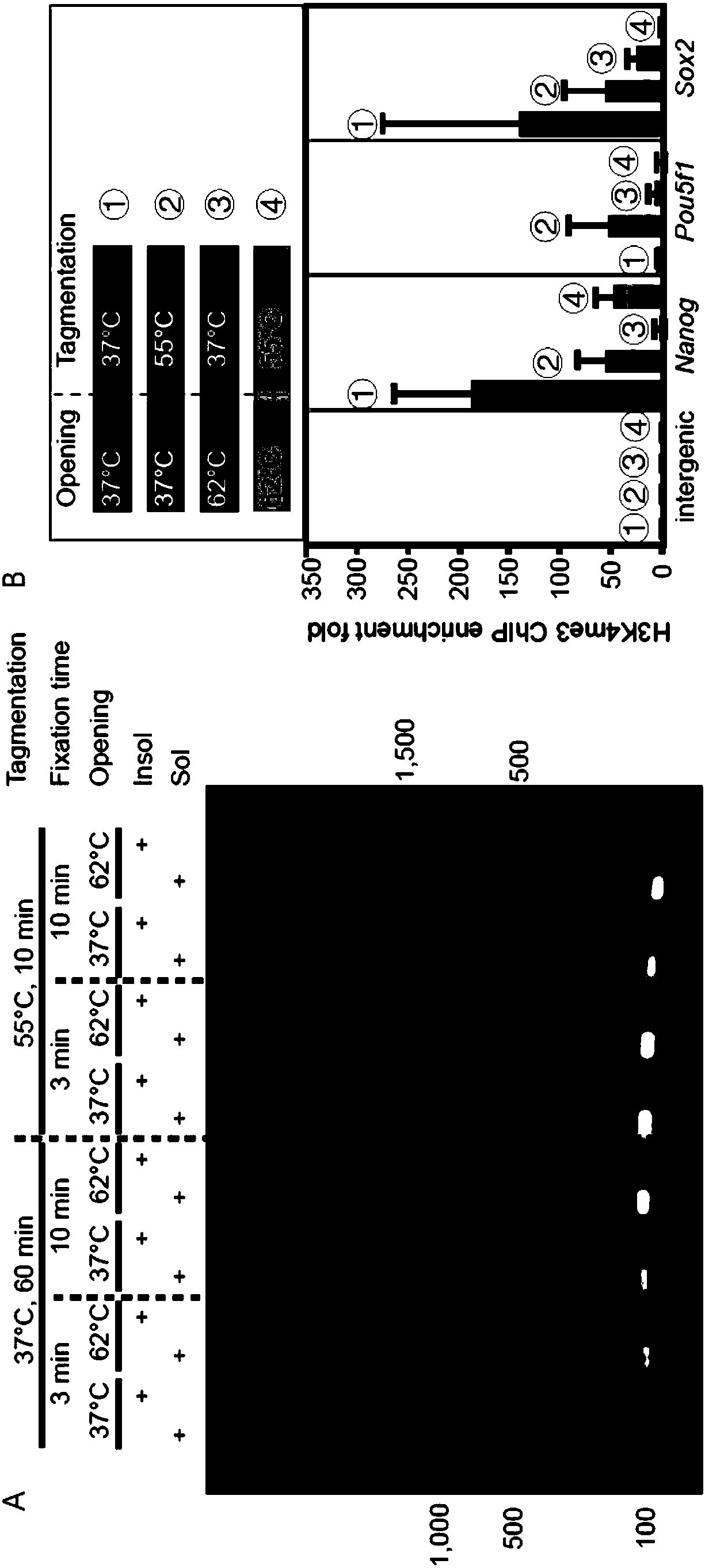
ActiveCN108315387AHigh catchImprove the efficiency of database constructionMicrobiological testing/measurementProtein targetEnzyme digestion
The invention relates to a trace cell ChIP (Chromatin Immunoprecipitation) method. The method comprises the following steps: 1) dividing a cell sample to be detected into n groups, wherein n is a natural number which is not equal to 0; 2) carrying out crosslinking fixation on the nth group sample; 3) treating the nth group sample by utilizing a cell membrane perforating agent; 4) carrying out enzyme digestion by utilizing Tn5 transposase and breaking a chromatin segment of the nth group sample, and connecting a barcode sequence and a primer sequence to two ends of a production segment DNA (Deoxyribonucleic Acid), wherein when the n is greater than or equal to 2, the barcode sequence and / or the primer sequence used in each group of step 4) are / is different; 5) when the n is greater than orequal to 2, combining samples of all groups; 6) enriching a DNA segment combined with target protein and taking a primer as an index for creating a database; sequencing and analyzing. When the methodis used for sequencing, the database creating efficiency is high; the capturing of transcription factor sites of an extremely trace amount of cells can be realized; relatively good efficiency is ensured and the stability and the accuracy are relatively good.
Owner:PEKING UNIV
Promoter batch capture method
ActiveCN107091929AEfficient separationEfficient acquisitionMicrobiological testing/measurementBiological testingConserved sequenceTATA box
The invention provides a promoter batch capture method. According to the promoter batch capture method, when a plant gene is transcribed, a complex of a 'DNA-protein factor' formed in a promoter area is utilized, an improved chromatin immunoprecipitation technology is applied, and DNA containing an unknown plant gene promoter fragment is sorted out. The promoter batch capture method is a technology for specifically capturing an unknown plant gene promoter fragment by utilizing the property that protein factors of OsTBP2 and OsTFIIB are tightly combined with promotion and transcribe of TATA box short-chain conserved sequence which on the promoter and through the generation of a specific antibody of the two factors and through the combination of the chromatin immunoprecipitation technology (ChIP). In the technology, by utilizing the advantages that most promoters contain one TATA box short-chain conserved sequence, and through the combination of technology that the unknown plant gene promoter fragment is separated through the newly developed chromatin immunoprecipitation technology (ChIP), large-scale capture of the plant promoter is easily achieved, and the technology has high commercial value.
Owner:RICE RES ISTITUTE ANHUI ACAD OF AGRI SCI
Ultrasonication method for heterocephalus glaber tissues in ChIP (Chromatin Immunoprecipitation)
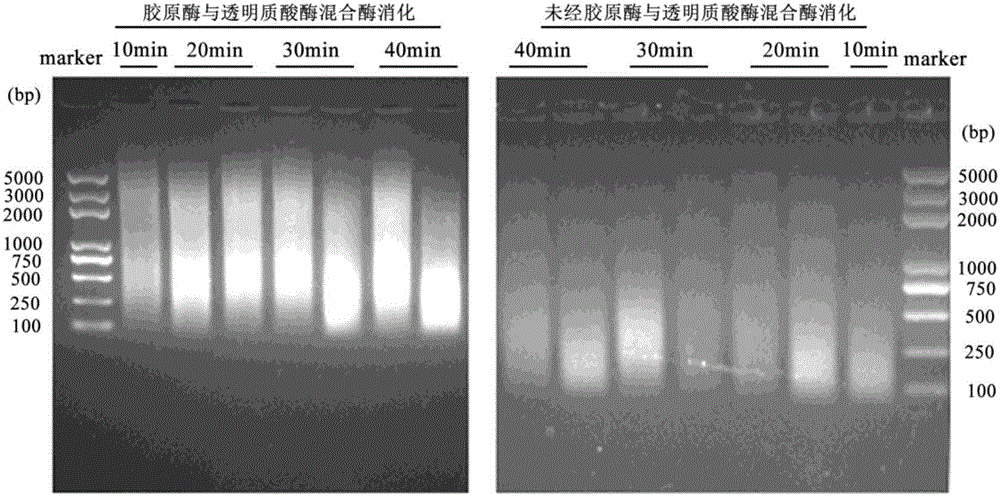
ActiveCN106834208AEasy accessAddress the shortcomings of poor performanceCell dissociation methodsMicroorganism lysisA-DNAProtein-protein complex
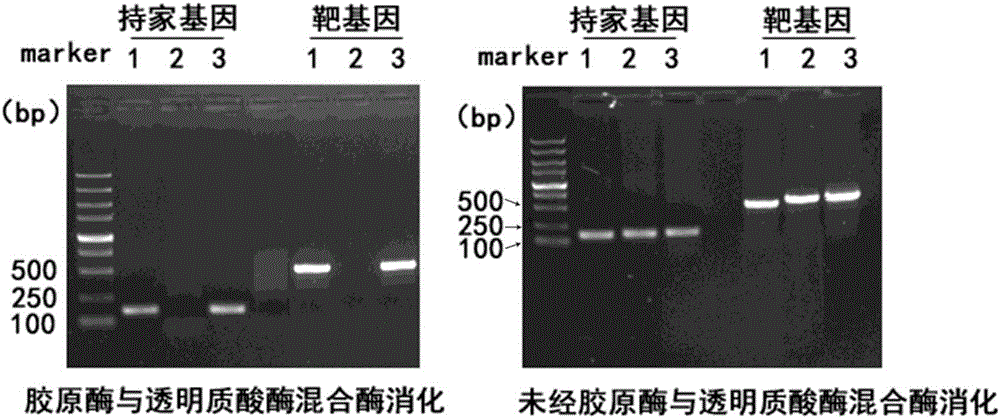
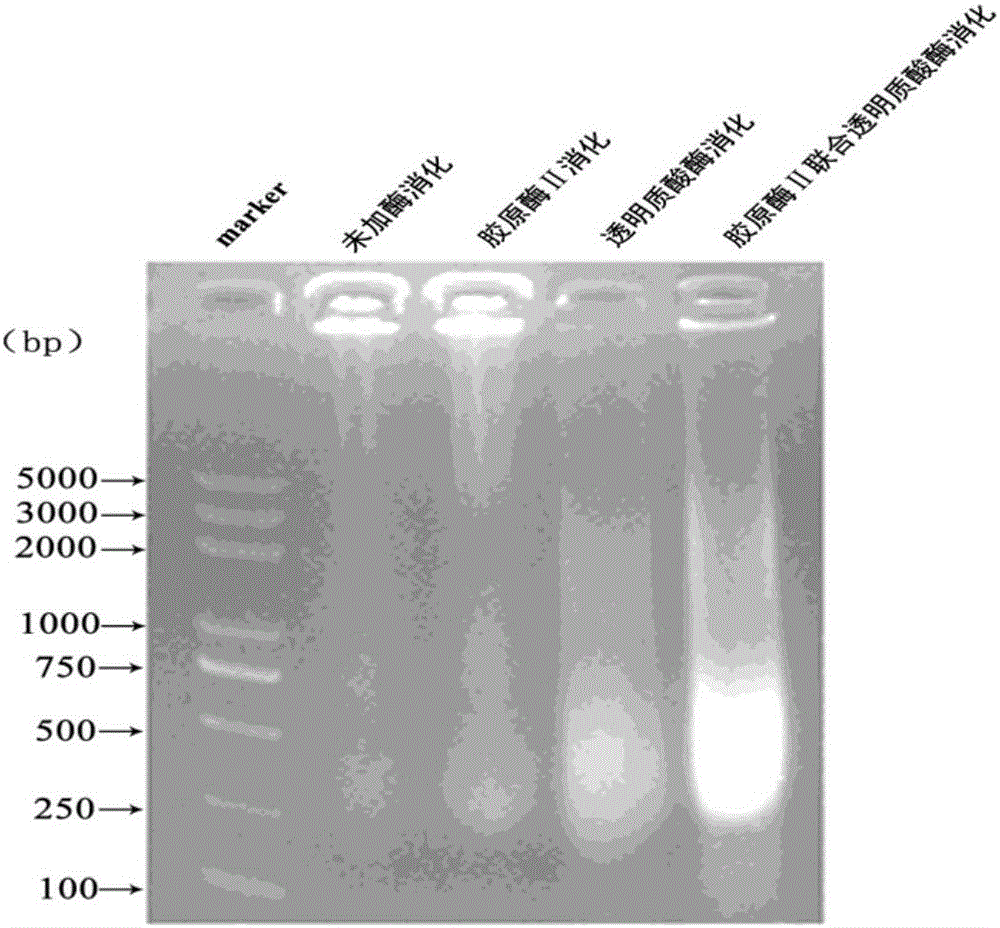
The invention relates to the field of immunoprcipitation, and in particular to an ultrasonication method for heterocephalus glaber tissues in ChIP (Chromatin Immunoprecipitation). The ultrasonication method comprises the following steps: firstly adding a mixed solution of a collagenase II solution, a hyaluronidase solution and a DPBS (Dulbecco's Phosphate Buffered Saline) buffer solution which are 1 to 1 to 1 in a volume ratio in the heterocephalus glaber tissues, oscillating under 37 DEG C for digesting tissues, centrifuging and removing supernatant, and then carrying out the next conventional ultrasonication step. By using the ultrasonication method disclosed by the invention, a DNA (Deoxyribonucleic Acid)-protein complex which is proper in fragment size can be well obtained, and a good foundation is laid for success of a whole follow-up ChIP experiment.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Data processing method for chromatin immunoprecipitation high-throughput sequencing
InactiveCN103853936AImprove work efficiencyReflect the distribution characteristicsSpecial data processing applicationsText fileThroughput
The invention relates to a data processing method for chromatin immunoprecipitation high-throughput sequencing, and belongs to the technical field of molecular biology. The method comprises the following steps: firstly eliminating low quality sequence data in an initial sequence file, then contrasting the filtered sequence data in a reference genome, counting signal peak amount and density distribution in different areas according to the classification of the reference genome, and determining neighboring genes of each signal peak for gene body function enrichment analysis, and finally generating a gene body function enrichment result text file and a corresponding graphical representation file. The method provides a high-efficiency and high-throughput data analysis process, so that each sequencing process is effectively integrated so as to help scientific research personnel to efficiently complete earlier-stage sequence quality control and sequence filtering of high-throughput data and reflect the advantages and disadvantages of a chromatin immunoprecipitation high-throughput sequencing experiment based on data statistics of contrasted sequence, and the distribution characteristics of the sequence on chromosome can be reflected, thus the work efficiency of sequencing is greatly promoted.
Owner:FENGHE SHANGHAI INFORMATION TECH
Pig tissue chromatin immunoprecipitation processing method
InactiveCN107254512AEasy to operatePromote resultsMicrobiological testing/measurementBiotechnologyEpigenome
The invention belongs to the technical field of livestock immunology, and concretely relates to a pig tissue chromatin immunoprecipitation processing method. The method is named as ChIP for short. The method specially aims at pig tissue for optimization design, employs a tissue lysate and a Dounce grinder for combination, and develops the method, the processes and the flows of pig tissue homogenate, the tissue can be fully cracked, the obtained chromatin amount is large, the quality is high, and the quality of a peak graph of the subsequent ChIP-Seq experiments is high. The method has the advantages of simpleness and rapidity, can effectively prepare a tissue suspension, can satisfy the core requirement of the ChIP-Seq experiments, and provides the effective approach for developing the pig tissue epigenomics researches.
Owner:HUAZHONG AGRI UNIV
Brachypodium distachyon functional centromere antigen polypeptide and application
InactiveCN105461789ADiscuss formationDiscuss functionSerum immunoglobulinsImmunoglobulins against plantsNew Zealand white rabbitNucleotide
The invention provides a brachypodium distachyon functional centromere histone CENH3 and application. Firstly, the nucleotide sequence of a brachypodium distachyon functional centromere histone CENH3 gene is predicted through the bioinformatics sequence comparison method, a polypeptide is synthesized according to a segment of specific sequence of the N end of an amino acid sequence encoded by the nucleotide sequence, New Zealand white rabbits are conventionally immuned through the polypeptide, antiserums are prepared, and a CENH3 protein antibody is obtained through antigen affinity purification. The CENH3 antibody can be used for immunostaining experiments, physical positioning of centromere and chromatin immunoprecipitation (ChIP) experiments, the functional DNA sequence of a brachypodium distachyon functional centromere is obtained, positioning of centromere genome genetic maps and physical maps is carried out, and important tools and information are provided for research of centromere formation and function exercise mechanisms and building of artificial chromosomes.
Owner:FUJIAN AGRI & FORESTRY UNIV
Identification method of BPDE adduct gene
The invention relates to the field of identification of BPDE adduct genes, in particular to an identification method of a BPDE adduct gene. Based on the principle of chromatin immunoprecipitation (ChIP), the BPDE-DNA adduct enriched by Anti-BPDE antibody is sequenced with high throughput to construct B [a] P adduct target gene site accurate localization technique. According to the method, the detected BPDE adduct target gene is a direct action between the BPDE and the gene, and small molecules do not need to be chemically modified in the detection process, so that the original activity of thesmall molecules is not influenced, the method is not limited by any cell and tissue type, pure DNA is not required to be used, even a whole cell lysate can be used, and the method can be widely applied to the identification of the small molecule adduct target gene.
Owner:SHANXI MEDICAL UNIV
Pineapple function centromere antigenic polypeptide and application thereof
InactiveCN105418740ADiscuss formationDiscuss functionSerum immunoglobulinsImmunoglobulins against plantsImmunofluorescenceNew Zealand white rabbit
The invention provides pineapple function centromere antigenic polypeptide and application thereof, an amino acid sequence is as shown in SEQ ID NO.2, one polypeptide is synthesized according to a specific sequence at an N terminal of the amino acid sequence, a New Zealand white rabbit is conventionally immunized by using the polypeptide, antiserum is prepared, an antibody of CENH3 protein is obtained by affinity purification of an antigen, an immunofluorescence test of the antibody is performed, the antibody of the pineapple CENH3 protein can be used for cytological localization of position of pineapple function centromere, also can be used for ChIP-seq combined with a chromatin immunoprecipitation technology of the pineapple, the position, size and sequence information of function centromere on chromosome of the pineapple are determined, the location of a centromere genome genetic map and a physical map is performed, and important tools and information are provided for formation of the centromere and study of a function performing mechanism and the construction of artificial chromosome.
Owner:FUJIAN AGRI & FORESTRY UNIV
Yunnan red pear delta PyTTG1 gene, prokaryotic expression vector thereof and application of prokaryotic expression vector
InactiveCN102586280AMicroorganism based processesImmunoglobulins against plantsEscherichia coliImmunoprecipitation
The invention discloses a Yunnan red pear pigment synthesis and trichome development regulatory protein PyTTG1 gene delta PyTTG1 and a prokaryotic expression vector thereof. The Yunnan red pear delta PyTTG1 purified protein is obtained by cloning the specific segment delta PyTTG1 of the PyTTG1 gene from the Yunnan red pear, constructing the prokaryotic expression vector of the gene and expressing the gene in the Escherichia coli. The Yunnan red pear delta PyTTG1 purified protein is applied to preparation of a PyTTG1 specific antibody, detection on PyTTG1 protein expression, immunoprecipitation, and chromatin immunoprecipitation. The antibody has high specificity and can accurately detect the subcellular localization of the PyTTG1 protein, efficiently purify the PtTTG1 protein, efficiently separate the protein bound with the PyTTG1 protein from the DNA segment, and detect the expression condition of the the PtTTG1 protein in transgenic plants.
Owner:KUNMING UNIV OF SCI & TECH
Pawpaw functional centromere antigen polypeptide, and applications thereof
InactiveCN105399805APromote formationFunction increaseSerum immunoglobulinsImmunoglobulins against plantsNew Zealand white rabbitImmunoprecipitation
The invention provides a pawpaw functional centromere antigen polypeptide, and applications thereof. The amino acid sequence of pawpaw functional centromere histone CENH3 is represented by SEQ ID No.2; a specific sequence of the N terminal of the amino acid sequence coded based on the pawpaw functional centromere histone CENH3 is used for synthesis of a polypeptide; routine immune-inoculation of New Zealand white rabbits with the polypeptide is carried out, antiserum is prepared, the antibody of CENH3 protein is obtained via antigen affinity purification, and immunofluorescence assay of the antibody is carried out. The antibody of CENH3 protein can be used for cellular localization of pawpaw functional centromere, and can be applied to pawpaw chromatin immunoprecipitation assay sequencing (ChIP-seq), so that the position, size, and sequence information of functional centromeres on pawpaw chromosomes can be determined, obtained physical map information is more complete, and centromere-related information can be provided for pawpaw genome assembly.
Owner:FUJIAN AGRI & FORESTRY UNIV
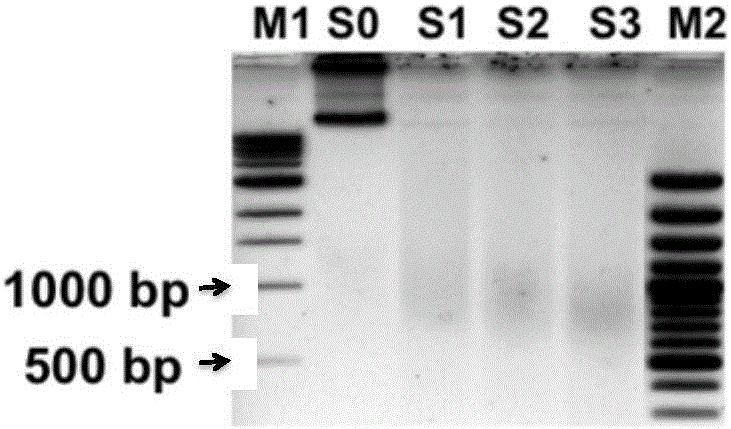
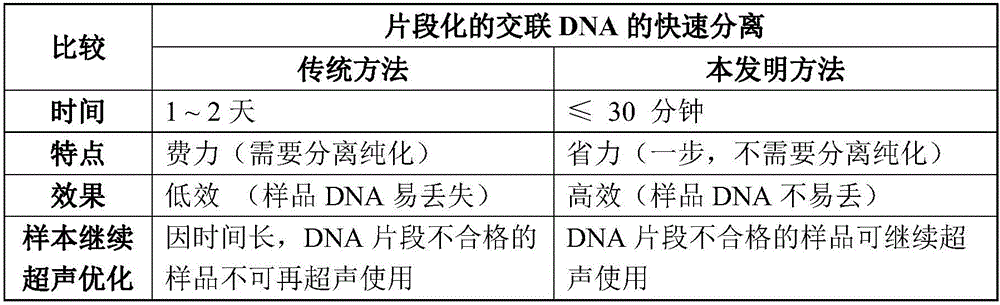
Rapid separation and detection kit and method for fragmented crosslinked DNA
InactiveCN105838783AIncrease success rateFast and effective quality control testingMicrobiological testing/measurementChelex 100A-DNA
The invention discloses a rapid separation and detection kit and method for fragmented crosslinked DNA. The kit includes a reagent for decrosslinking and separation extraction of fragmented DNA, and the reagent is Chelex 100. The kit can further include a protein degradation reagent, a DNA ladder marker, a sample loading buffer solution and / or an RNA degradation reagent. Chelex 100 can integrate multivalent metal ions, has high metal ion selectivity and binding force, can separate DNA and prevent DNA degradation. The rapid separation and detection method for fragmented crosslinked DNA can determine whether a to-be-detected sample needs further ultrasound or is used for a next step precipitation experiment according to the effect of rapid quality control ultrasound fragmentation, or can be used for ultrasound optimization of a plurality of to-be-detected samples so as to select optimum conditions. The kit and the method provided by the invention can be used for rapid detection of chromatin ultrasound fragmented sample quality in chromatin immunoprecipitation assay (ChIP) and other technologies, and can greatly improve the success rate of ChIP.
Owner:BEIJING JIAOTONG UNIV +1
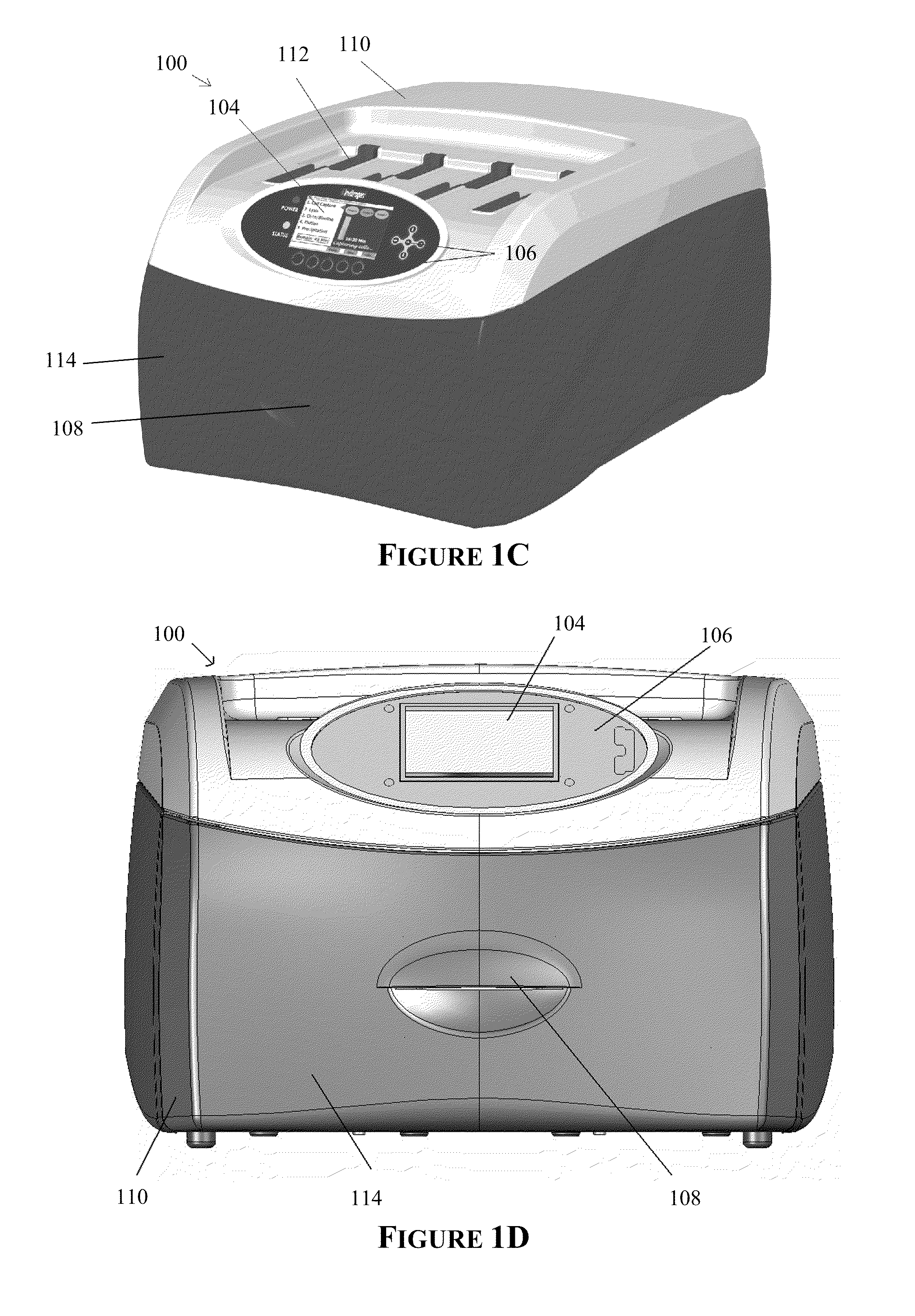
Apparatus for and method of processing biological samples
ActiveUS20200197934A1Heating or cooling apparatusLaboratory glasswaresBacterial virusProtein markers
The present invention provides systems, devices, apparatuses and methods for automated bioprocessing. Examples of protocols and bioprocessing procedures suitable for the present invention include but are not limited to: immunoprecipitation, chromatin immunoprecipitation, recombinant protein isolation, nucleic acid separation and isolation, protein labeling, separation and isolation, cell separation and isolation, food safety analysis and automatic bead based separation. In some embodiments, the invention provides automated systems, automated devices, automated cartridges and automated methods of western blot processing. Other embodiments include automated systems, automated devices, automated cartridges and automated methods for separation, preparation and purification of nucleic acids, such as DNA or RNA or fragments thereof, including plasmid DNA, genomic DNA, bacterial DNA, viral DNA and any other DNA, and for automated systems, automated devices, automated cartridges and automated methods for processing, separation and purification of proteins, peptides and the like.
Owner:LIFE TECH CORP
Methods for quantification of nucleosome modifications and mutations at genomic loci and clinical applications thereof
PendingCN111836907AMicrobiological testing/measurementOrganic compound librariesDiseaseEpigenetic Profile
The invention relates to clinical applications of quantitative chromatin mapping assays, such as chromatin immunoprecipitation assays and assays using tethered enzymes (e.g., chromatin immunocleavage(ChIC) and cleavage under targets & release using nuclease (CUT&RUN )). The methods may be used to detect and quantitate the presence of epigenetic modifications and mutations on nucleosomes (histonesand / or DNA) from biological samples, monitor changes in the status of modifications and mutations, monitor the effectiveness of epigenetic and mutation therapies, select suitable treatments for a disease, determine the prognosis of a subject, identify biomarkers of a disease, and screen for agents that modify epigenetic or mutation status. The invention further relates to kits for use in the methods of the invention.
Owner:埃皮赛佛尔有限公司
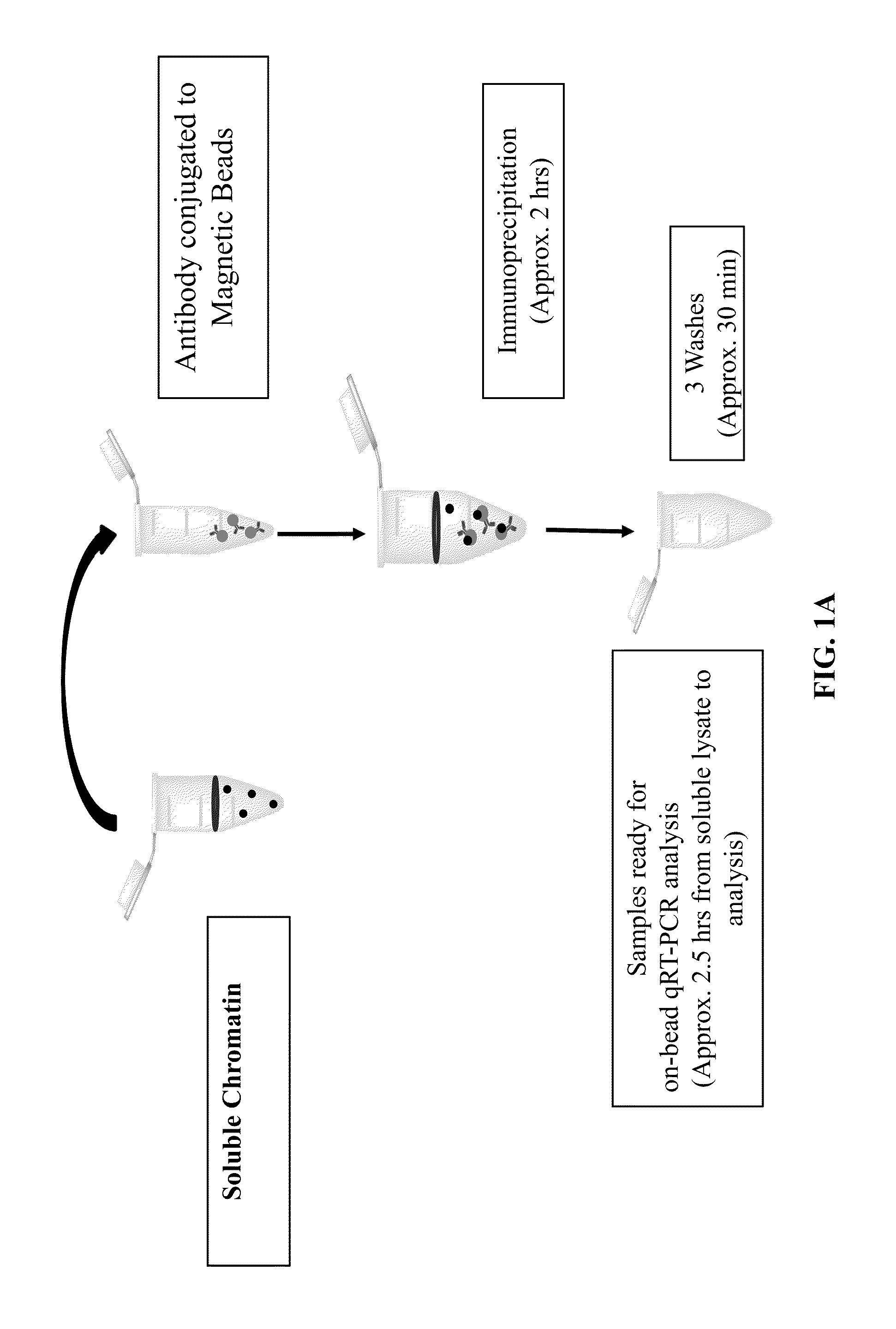
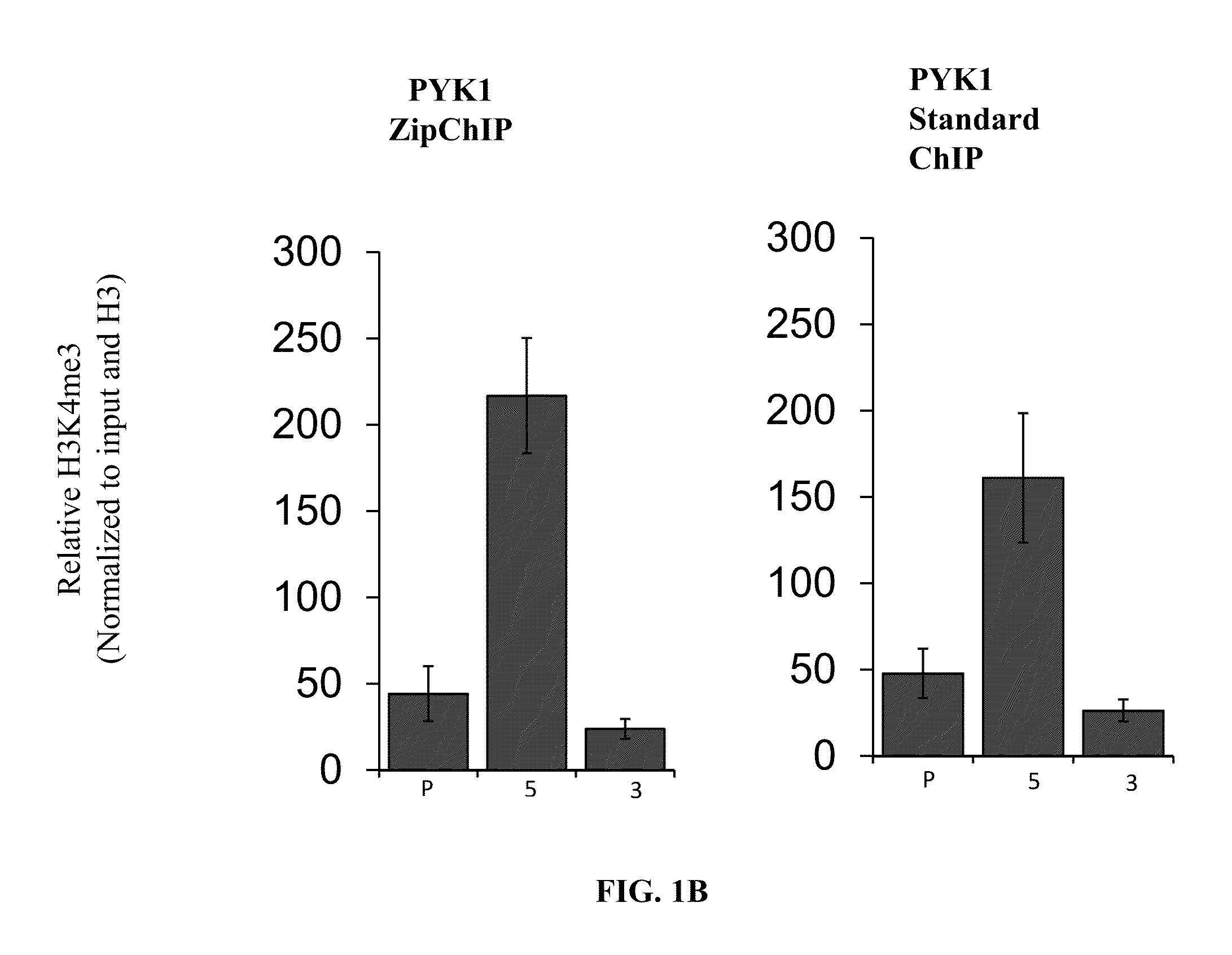
Immediate chromatin immunoprecipitation and analysis
InactiveUS20160168622A1Reduce the amount of solutionShorten the timePeptide librariesNucleotide librariesYeastHistone deacetylase
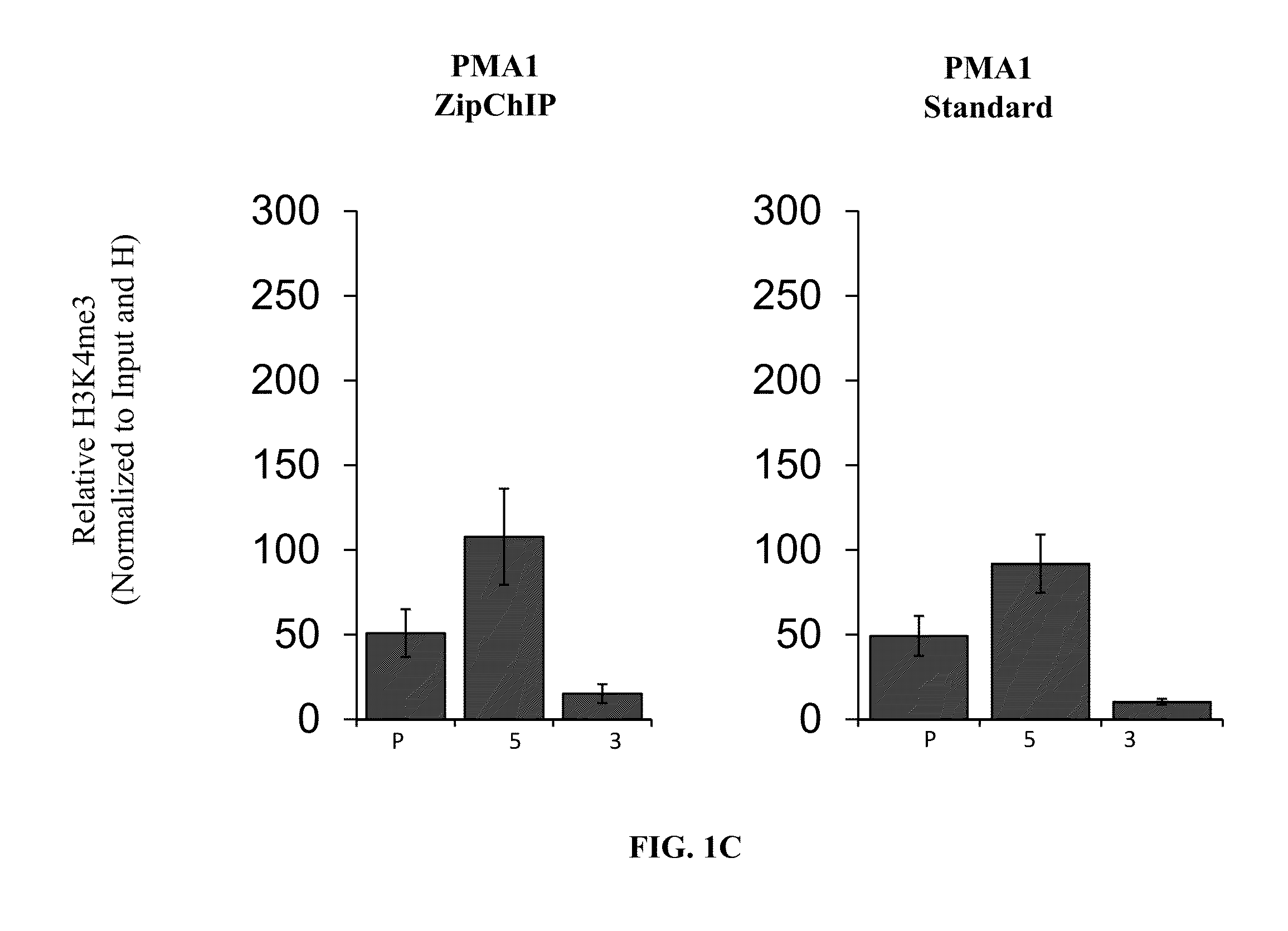
The present invention relates to a newly developed immediate chromatin immunoprecipitation procedure (“ZipChIP”). ZipChIP significantly reduces the time and increases sensitivity allowing for rapid screening of multiple loci. ZipChIP enables the detection of histone modifications (e.g., H3K4 mono- and trimethylation) and at least two yeast histone demethylases, Jhd2 and Rph1, which were previously found difficult to detect using standard methods. ZipChIP further relates to the enrichment of the histone deacetylase Sir2 at heterochromatin in yeast and enrichment of the chromatin remodeler, PICKLE, in Arabidopsis thaliana.
Owner:PURDUE RES FOUND INC
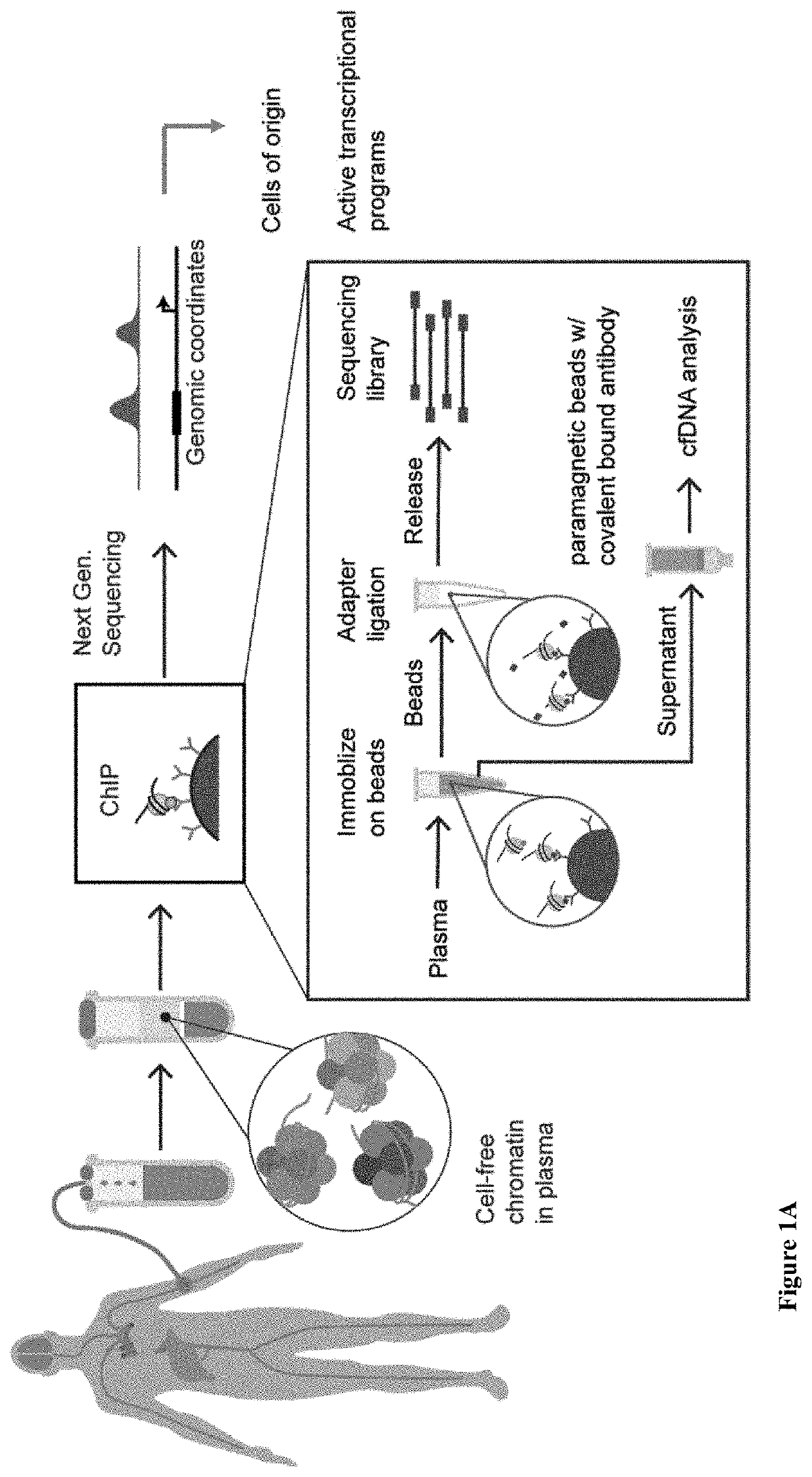
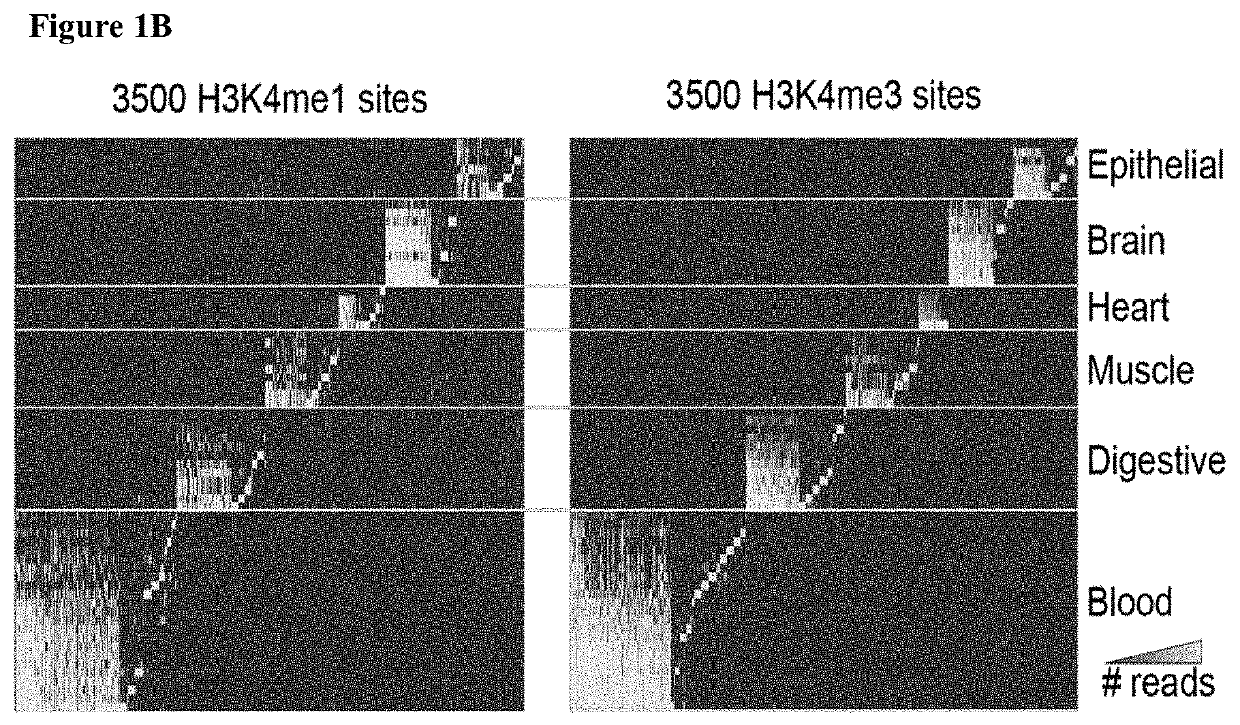
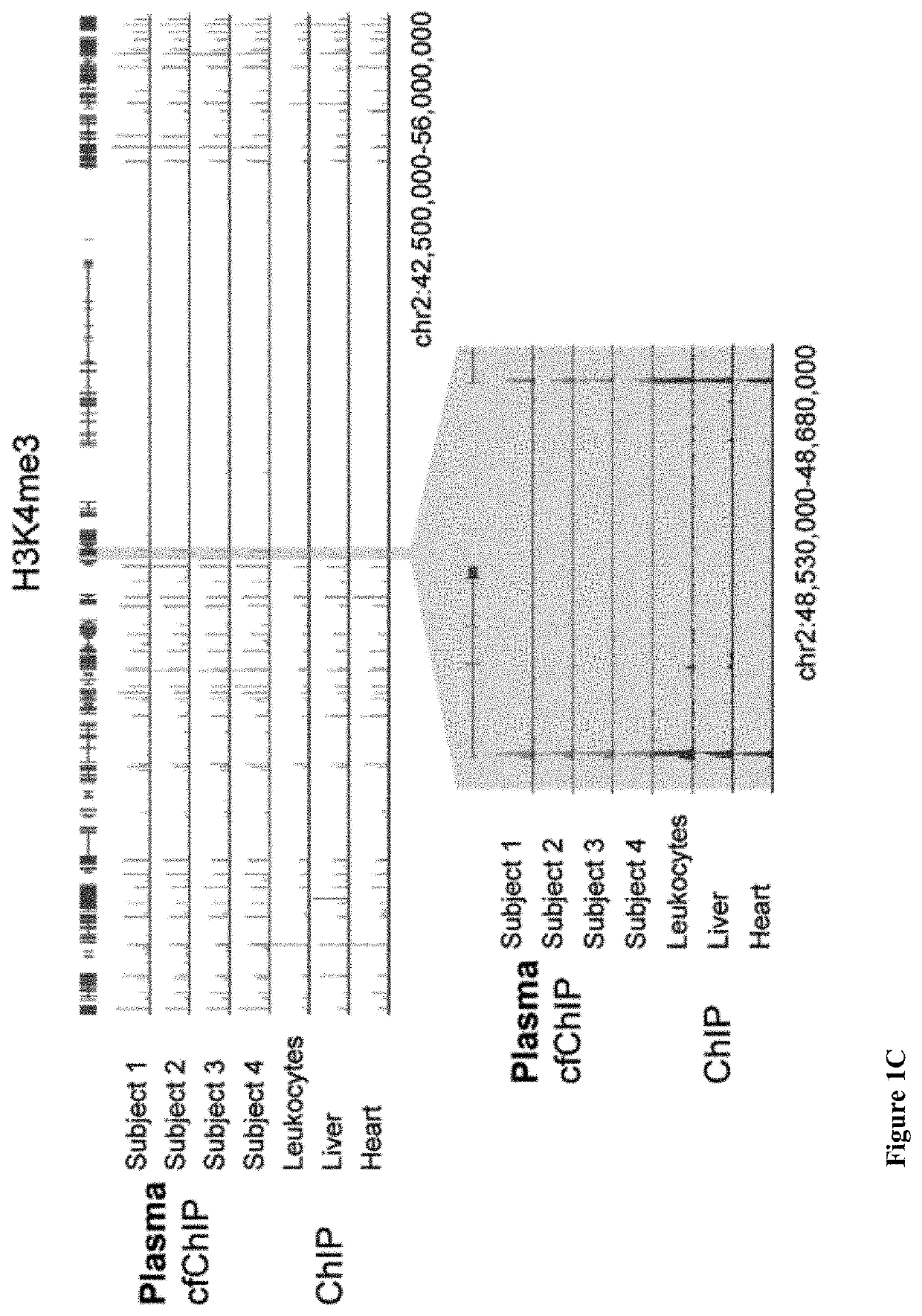
Diagnostic use of cell free DNA chromatin immunoprecipitation
Methods of determining the origin of cell free DNA (cfDNA), for detecting death of a cell type or tissue in a subject, for determining a cellular state of a cell as it died, and combinations thereof, are provided. As are computer program products for doing same.
Owner:YISSUM RES DEV CO OF THE HEBREWUNIVERSITY OF JERUSALEM LTD
Compositions and methods comprising the use of cell surface displayed homing endonucleases
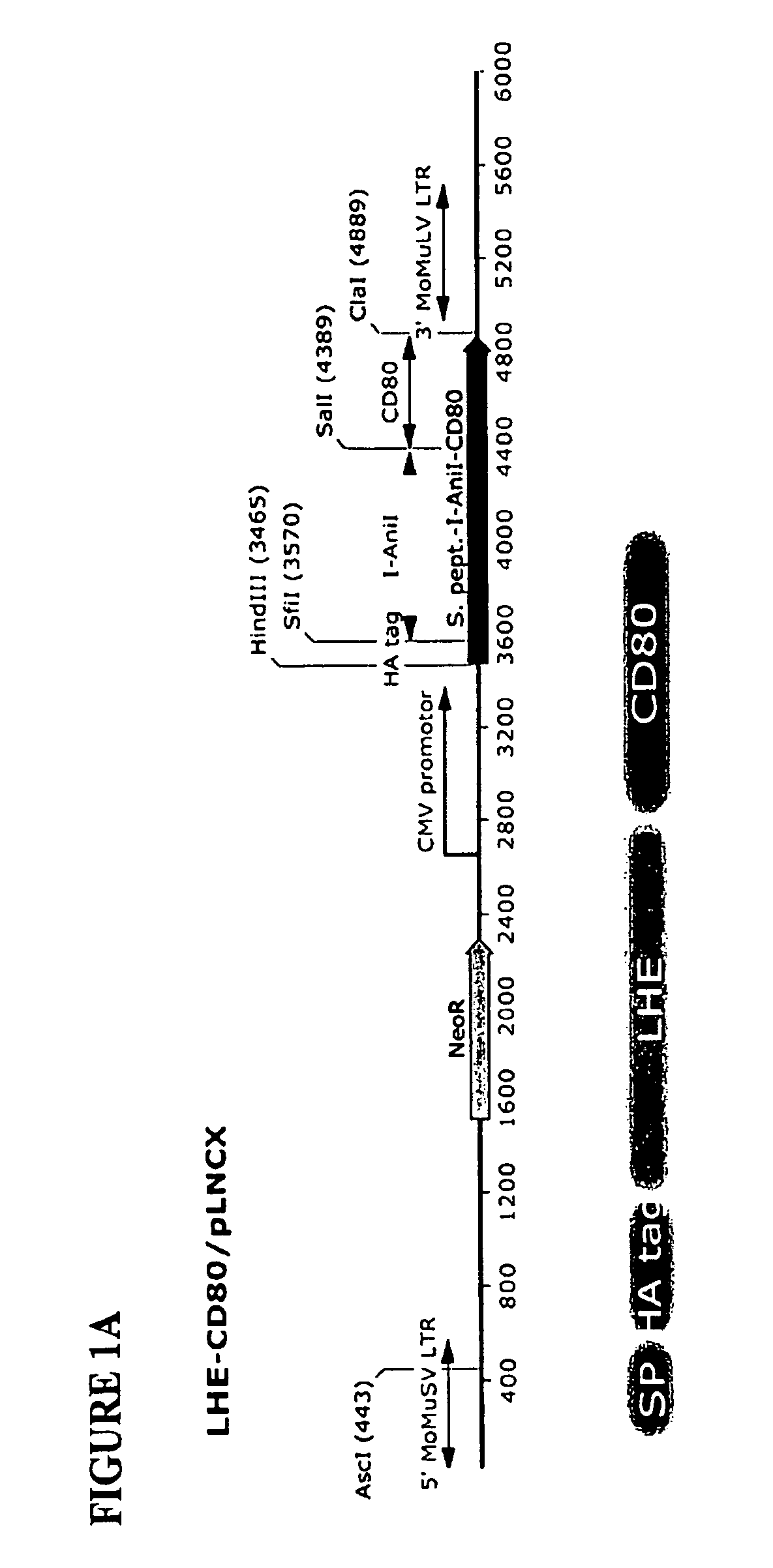
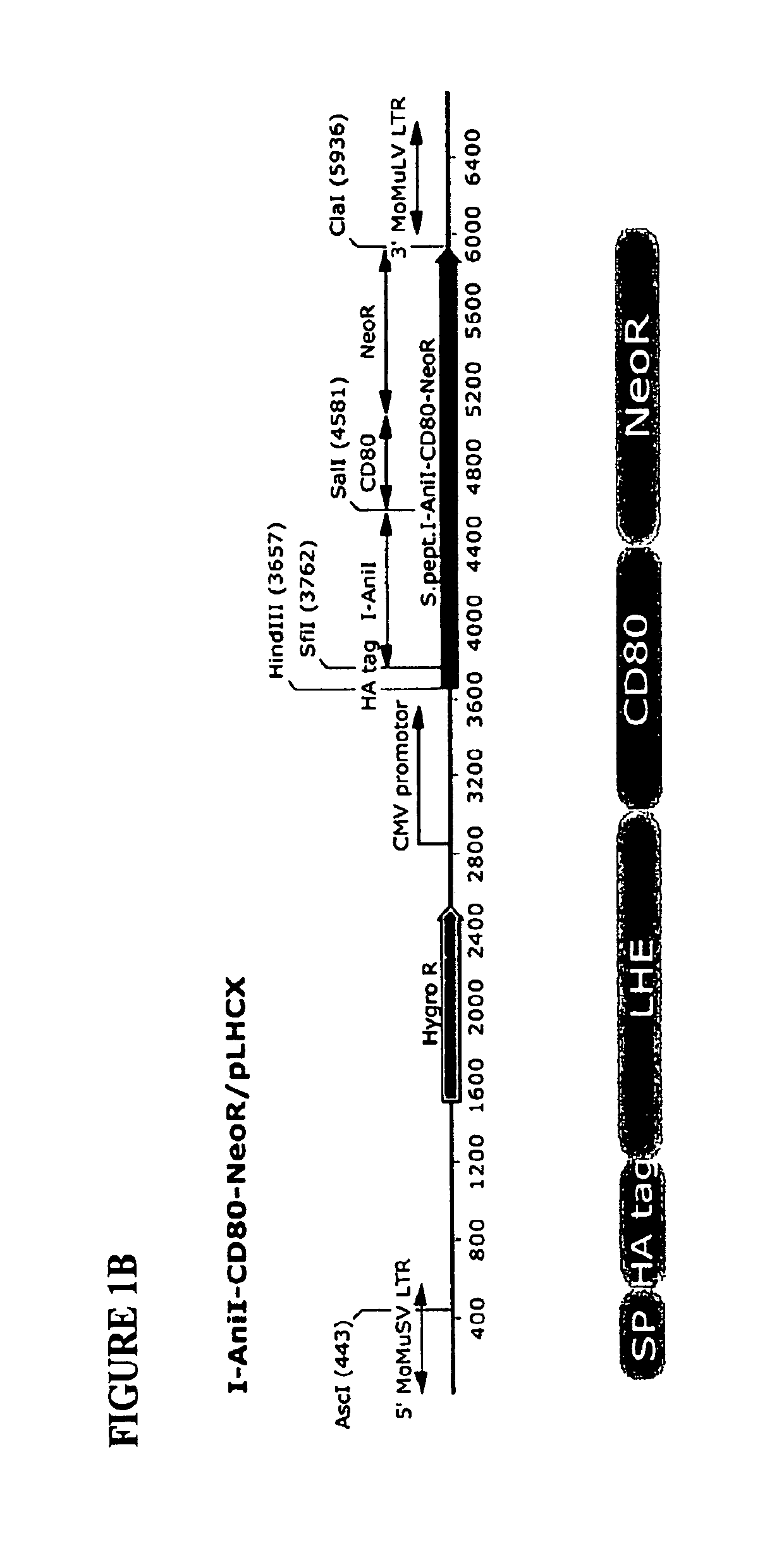
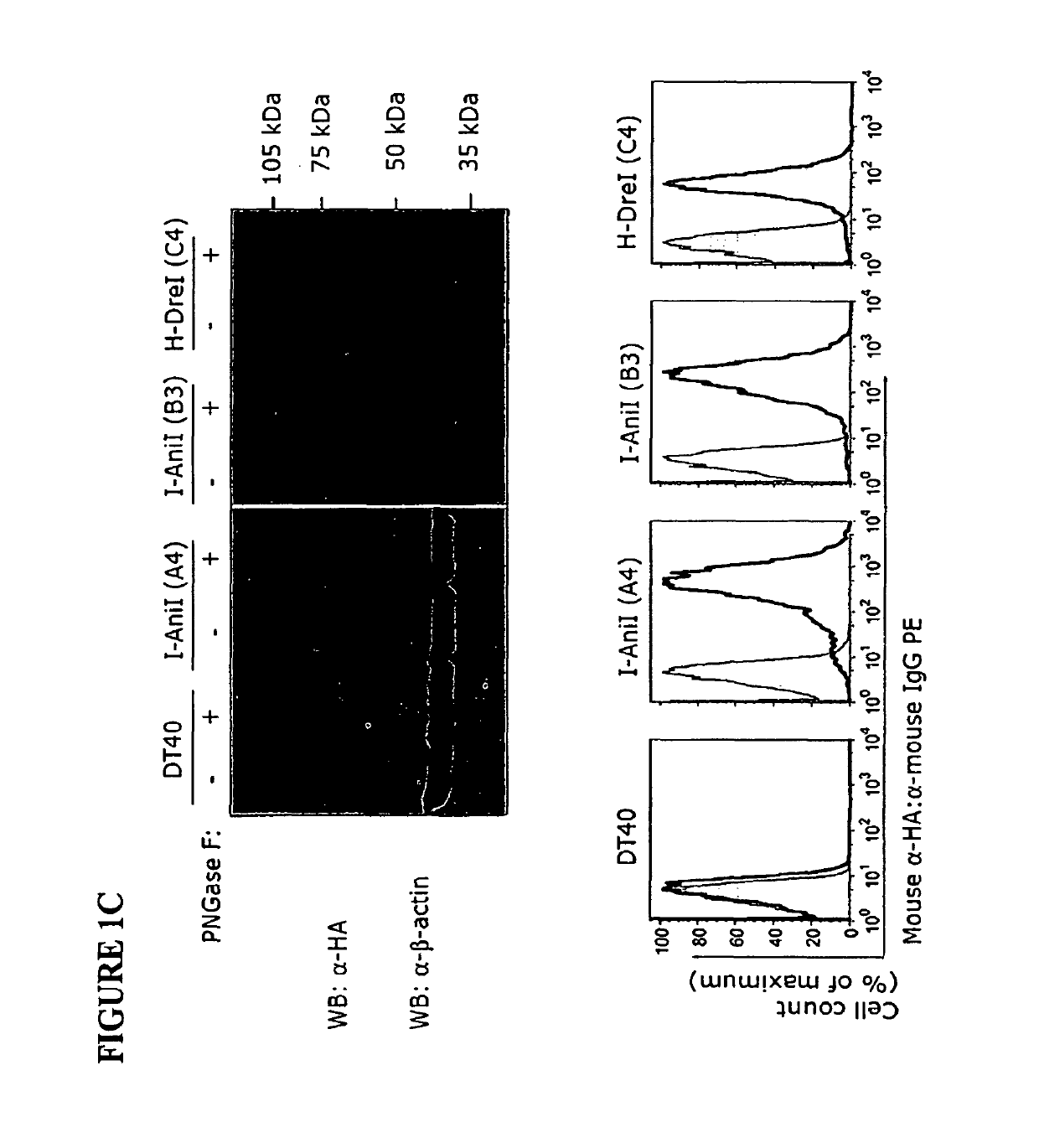
Owner:SEATTLE CHILDRENS HOSPITAL
Methods for preparing sequencing libraries
ActiveUS8574832B2Reduce the amount requiredSugar derivativesMicrobiological testing/measurementImproved methodHigh throughput sequence
Improvements in chromatin immunoprecipitation-high throughput sequencing techniques has allowed the creation of chromatin maps from limited biological sample sizes that cannot be evaluated using conventional chromatin immunoprecipitation-sequencing protocols. For example, a modified universal primer is utilized that incorporates restriction enzymes into chromatin immunoprecipitation fragments before amplification. The improved method allows the sample sizes to be several orders of magnitude less than that required for standard ChIP-Seq techniques.
Owner:THE GENERAL HOSPITAL CORP +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com