Method for detecting DNA of schistosoma through fluorescent probe
A fluorescent probe, schistosomiasis technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as false positives, long time consumption, contamination of amplification products, etc., and achieve simple operation, easy collection, and short time. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
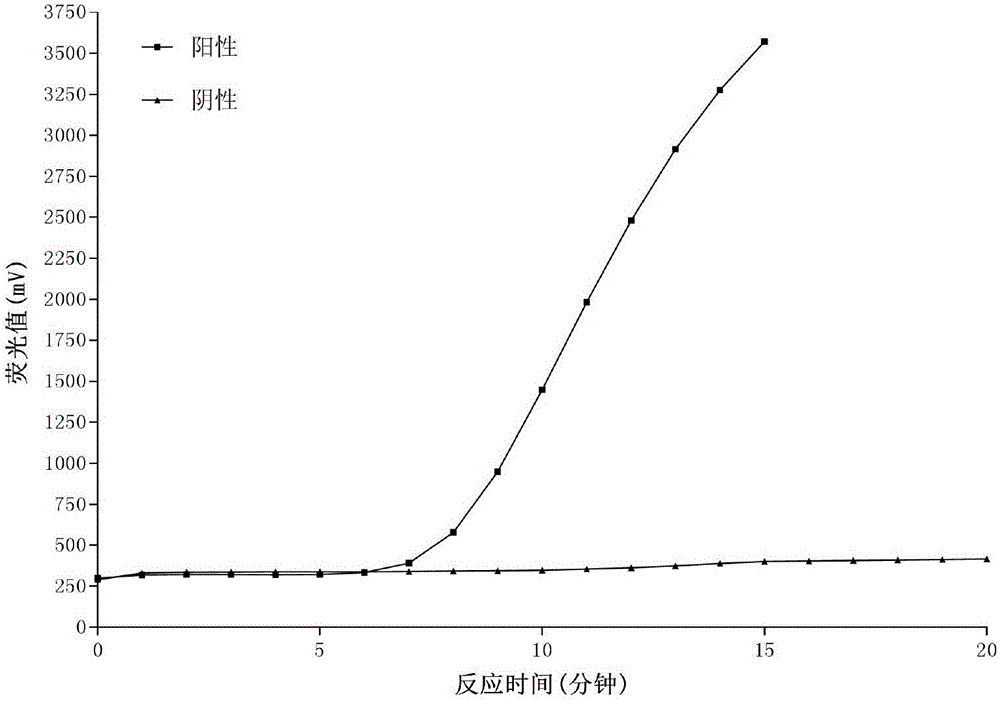
[0070] Sample source: Genomic DNA extracted from adults of Schistosoma japonicum, the concentration of DNA is 50ng / μL.
[0071] Design the following schistosome primers and probes, and entrust Sangon Bioengineering (Shanghai) Co., Ltd. to synthesize them:
[0072] Forward primer sequence: 5'-TACCTCAAGAAGTAATGTCCTTCCATTGTG-3',
[0073] Reverse primer sequence: 5'-ATGCGAGGTTTCAGGAGACCAAGAAGAACG-3'.
[0074] The sequence of the fluorescent probe is:
[0075] 5'CATAGGAGGTCATCTTGTTCAAGGTCAAGTCTCACCATCAACTCTTA-3'
[0076] Prepare the amplification system
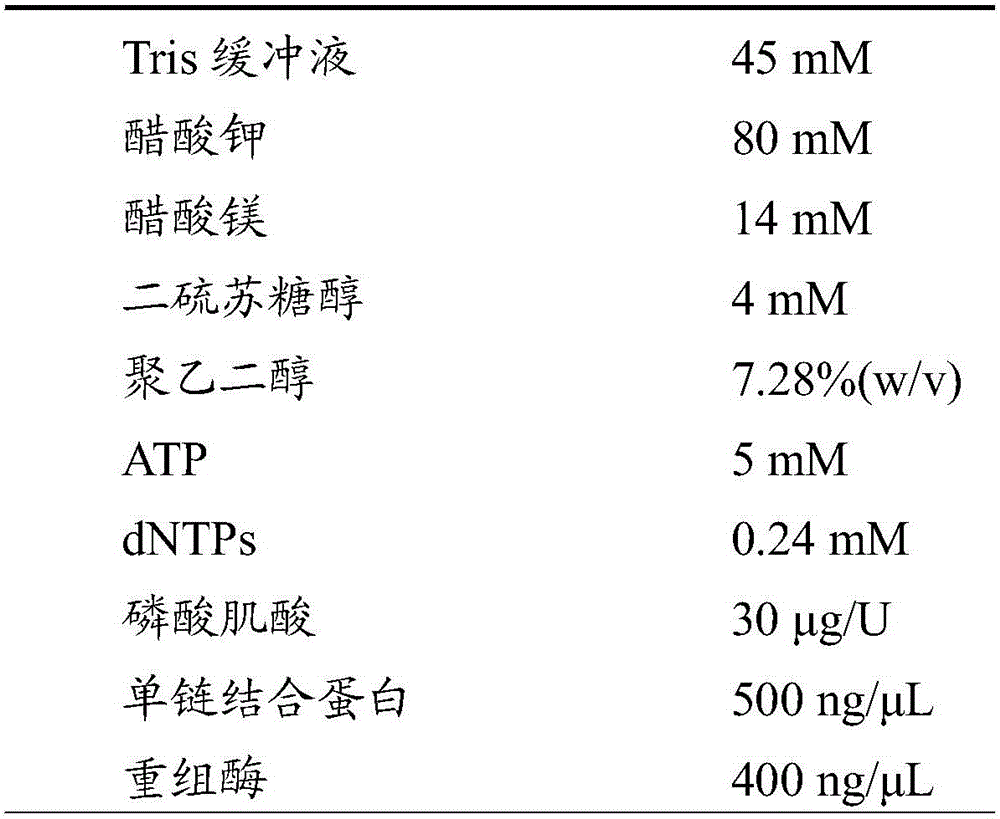
[0077] Prepare the isothermal nucleic acid amplification system (volume 50 μL) in a 200 μL centrifuge tube according to the following ratio:
[0078]
[0079]
[0080] The above-prepared amplification system was lyophilized under negative pressure in a freeze dryer to become a powdery amplification system.
[0081] Add polyethylene glycol with a final concentration of 6% (w / v) and a molecular weight of 35,000 to the c...
Embodiment 2
[0083] Sample source: Genomic DNA extracted from adults of Schistosoma japonicum, the concentration of DNA is 70ng / μL.
[0084] Design the following schistosome primers and probes, and entrust Sangon Bioengineering (Shanghai) Co., Ltd. to synthesize them:
[0085] Forward primer sequence: 5'-TACCTCAAGAAGTAATGTCCTTCCATTGTG-3',
[0086] Reverse primer sequence: 5'-ATGCGAGGTTTCAGGAGACCAAGAAGAACG-3'.
[0087] The sequence of the fluorescent probe is:
[0088] 5'CATAGGAGGTCATCTTGTTCAAGGTCAAGTCTCACCATCAACTCTTA-3'
[0089] Prepare the amplification system
[0090] Prepare the isothermal nucleic acid amplification system (volume 100 μL) in a 200 μL centrifuge tube according to the following ratio:
[0091]
[0092]
[0093] The above-prepared amplification system was lyophilized under negative pressure in a freeze dryer to become a powdery amplification system.
[0094] Add polyethylene glycol with a final concentration of 6% (w / v) and a molecular weight of 35,000 to the ...
Embodiment 3
[0096] Sample source: Genomic DNA extracted from adults of Schistosoma japonicum, the concentration of DNA is 82ng / μL.
[0097] Design the following schistosome primers and probes, and entrust Sangon Bioengineering (Shanghai) Co., Ltd. to synthesize them:
[0098] Forward primer sequence: 5'-TACCTCAAGAAGTAATGTCCTTCCATTGTG-3',
[0099] Reverse primer sequence: 5'-ATGCGAGGTTTCAGGAGACCAAGAAGAACG-3'.
[0100] The sequence of the fluorescent probe is:
[0101] 5'CATAGGAGGTCATCTTGTTCAAGGTCAAGTCTCACCATCAACTCTTA-3'
[0102] Prepare the amplification system
[0103] Prepare the isothermal nucleic acid amplification system (volume 50 μL) in a 200 μL centrifuge tube according to the following ratio:
[0104]
[0105] The above-prepared amplification system was lyophilized under negative pressure in a freeze dryer to become a powdery amplification system.
[0106] Add polyethylene glycol with a final concentration of 6% (w / v) and a molecular weight of 35,000 to the centrifuge tu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com