cDNA specific molecular tag and application thereof
A technology of molecular labeling and specificity, applied in the biological field, can solve the problems of inaccuracy and the inability of biological information analysis data to truly reflect gene expression, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Design of specific molecular tags for cDNA
[0030] Specific molecular tags for cDNA include:
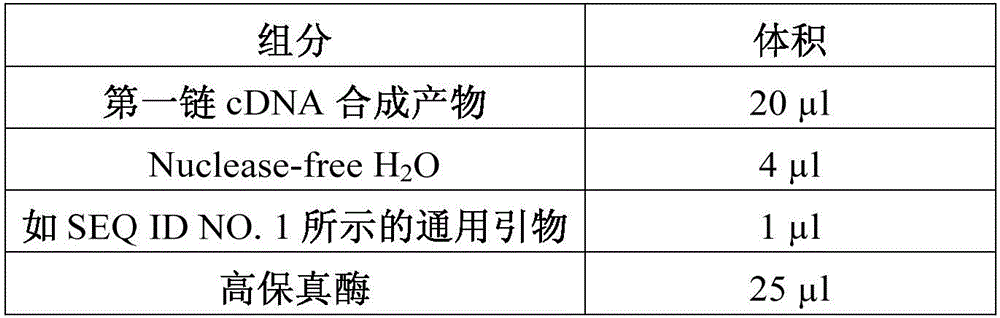
[0031] A. The universal primer sequence, as shown in SEQ ID NO.1, is used for subsequent PCR amplification of full-length cDNA. The specific sequence of the universal primer is as follows: AAGCAGTGGTATCAACGCAGAGTAC;
[0032] B. Transposase linker sequence P7, as shown in SEQ ID NO.2, the specific sequence is as follows: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG;
[0033] C. Random sequence (N) n , where N is a random sequence, a total of n;
[0034] D, poly(T) sequence, a total of 30 T bases, namely T30VN;
[0035] A cDNA molecular tag was synthesized by a gene synthesis method, and its nucleotide sequence is shown in SEQ ID NO.3: AAGCAGTGGTATCAACGCAGAGTAC GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG(N) n T30VN, wherein, N is a random sequence, a total of n, N is selected from A, T, C, G; preferably, n is 5-10.
Embodiment 2
[0036] Example 2 Preparation of full-length cDNA with the specific molecular tag
[0037] Human 293 cells were selected as the experimental material, and transcriptome amplification was performed using the Discover-sc WTA KitV2 (N711) kit from Vazyme. The reagents and materials used are provided by the kit, for example, single cell lysate, RNase Inhibitor, reverse transcription buffer, DTT, TSOligo, reverse transcriptase, high-fidelity enzyme, etc.
[0038] (1) Sample preparation
[0039] 1) Prepare 10×Sample Buffer in the following order, mix gently with a pipette, and collect by short centrifugation.
[0040] Table 1
[0041] components volume single cell lysate 19μl RNase Inhibitor 1μl total capacity 20μl
[0042] 2) Take 1 μl of 10×Sample Buffer into a 0.2ml PCR tube, then add 4 μl of nuclease-free pure water (Nuclease-free H 2 O).
[0043] 3) Isolation of single cells
[0044] Take the freshly cultured 293 cell suspension, collect...
Embodiment 3
[0069] Example 3 DNA library construction method using transposase
[0070] Use the (Vazyme) TruePrep DNA Library PrepKit V2 for Illumina (TD503) kit produced by Nanjing Nuoweizan Biotechnology Co., Ltd., and the reagents and materials used are all provided by the kit, for example, transposase buffer, PCR buffers, high-fidelity enzymes, and more.
[0071] The transposase and embedding buffer were provided by (Vazyme) Tn5Transposome (S111) kit produced by Nanjing Novizan Biotechnology Co., Ltd.
[0072] (1) Joint embedding
[0073] Configure the reaction system shown in Table 7, gently blow and mix with a pipette, place the prepared reaction system on a PCR instrument at 30°C for 1.5 hours to obtain the transposase embedding complex, and store at -20°C .
[0074] Table 7
[0075]
[0076]
[0077] (2) Fragmentation of full-length cDNA with molecular tags
[0078] 1) Add each reaction component shown in Table 8 in sequence to a sterilized PCR tube, and gently blow and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com