Mutant ubiquitination-specific protease 33 gene and its application
A specific, protease technology, applied in the field of molecular biology, which can solve the problems of inability to deubiquitinate Robo receptors in time, low expression, tumor cell invasion and migration, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
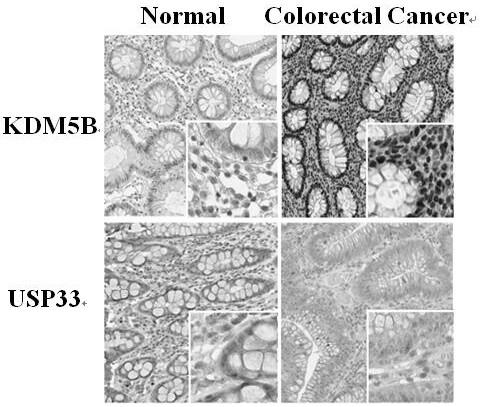
[0020] Example 1 KDM5B is highly expressed in colorectal cancer cells and down-regulates the expression of USP33
[0021] Routine immunohistochemical analysis was performed on 273 clinical cases of colorectal cancer tissue samples. The results showed that KDM5B was significantly highly expressed in colorectal cancer tissues, while USP33 was significantly less expressed, as shown in figure 1 shown.
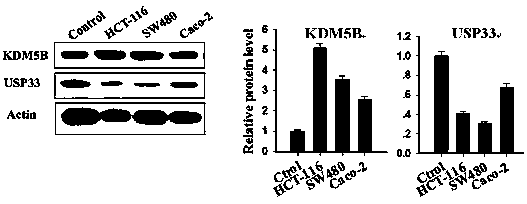
[0022] Conventional cell culture techniques were used to culture human colorectal cancer cell lines HCT-16, SW480, Caco-2 and normal human colon cells NCM460, and harvest the cells when the cells were growing well. Various cell lysates were obtained by conventional cell protein preparation technology, and the expression levels of KDM5B and USP33 in the above cells were detected by conventional Western-blot technology. The results showed that the expression of KDM5B and USP33 in colon cancer cells was negatively correlated, as shown in figure 2 shown. Antibodies used in Western-...
Embodiment 2
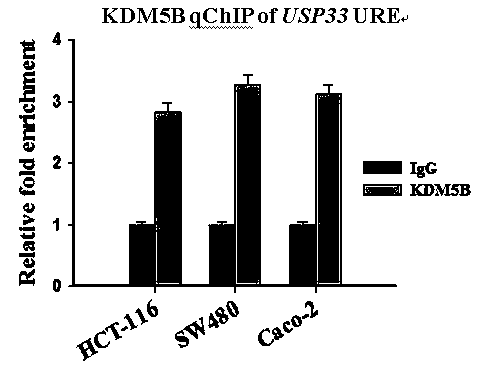
[0023] Example 2 KDM5B binds to the USP33 gene promoter region
[0024] To clarify whether KDM5B binds to the USP33 gene promoter region, a qChIP protocol was used. details as follows:
[0025] (1) Collect cultured colon cancer cells at room temperature, centrifuge at 2000 g for 5 min to collect 4×10 7 Cells were resuspended in 4°C pre-cooled cell culture medium, transferred to a 15ml centrifuge tube, and placed in ice for 10 min.
[0026] (2) Use 1% formaldehyde solution for cross-linking reaction, the purpose is to stabilize the combination of protein factors and DNA, and stop with 0.125M glycine after the cross-linking reaction.
[0027] (3) Centrifuge to obtain the cell pellet, first use 10ml of L1 solution (50 mMHepes KOH, pH7.5, 140 mMNaCl, 1 mM EDTA, 10% Glycerol, 0.5% NP-40, 0.25% Triton X-100) to lyse, 4°C, 10 min. Centrifuge after L1 lysis, 3000rpm, 10min, 4°C.
[0028] (4) The obtained cell pellet was further lysed with 10ml of L2 solution (0.2 M NaCl, 1 mM EDT...
Embodiment 3
[0041] Example 3 KDM5B down-regulates the transcription of USP33 gene
[0042] To confirm the effect of KDM5B on USP33 gene transcription, a luciferase reporter assay was used, as follows:
[0043] (1) Construction of the luciferase assay reporter vector. Get the upstream 3kb sequence of USP33 from the UCSC website (http: / / genome.ucsc.edu / ), design the upstream and downstream primers (see attached table) according to the genome sequence, get USP33-3kb, 2kb, 1kb promoter sequences respectively, and insert KpnI and XhoI restriction sites in the pGL4.15 vector were used to construct pGL4.15-USP33-3kb, pGL4.15-USP33-2kb and pGL4.15-USP33-1kb, respectively. The KDM5B expression vector was constructed, primers were designed according to the KDM5B gene (NM_006618.3), and then cloned into the pcDNA3.1-HA vector NheI and
[0044]
[0045] (2) Cell transfection test. Using conventional cell transfection techniques, the above constructed pcDNA3.1-KDM5B and pGL4.15-USP33-3kb, or pGL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com