Protein isotope dilution tandem mass spectrometry method based on chemical labeling technology
A technology of isotope dilution and tandem mass spectrometry, applied in the field of biological detection, can solve the problems of high price, cumbersome operation, and inability to correct errors, and achieve the effect of wide application range, wide selection range and convenient operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
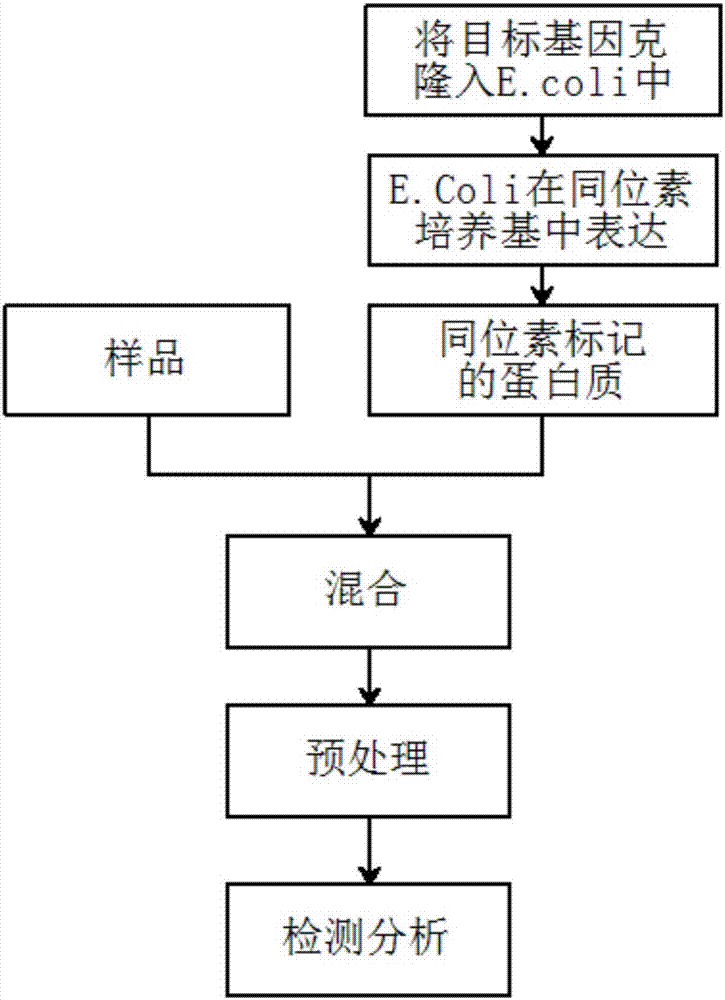
[0038] This embodiment is the detection of alpha s1-casein (milk allergen) in red wine, wherein the alpha s1-casein content is 20 μg / mL, and the detection process includes the following steps:
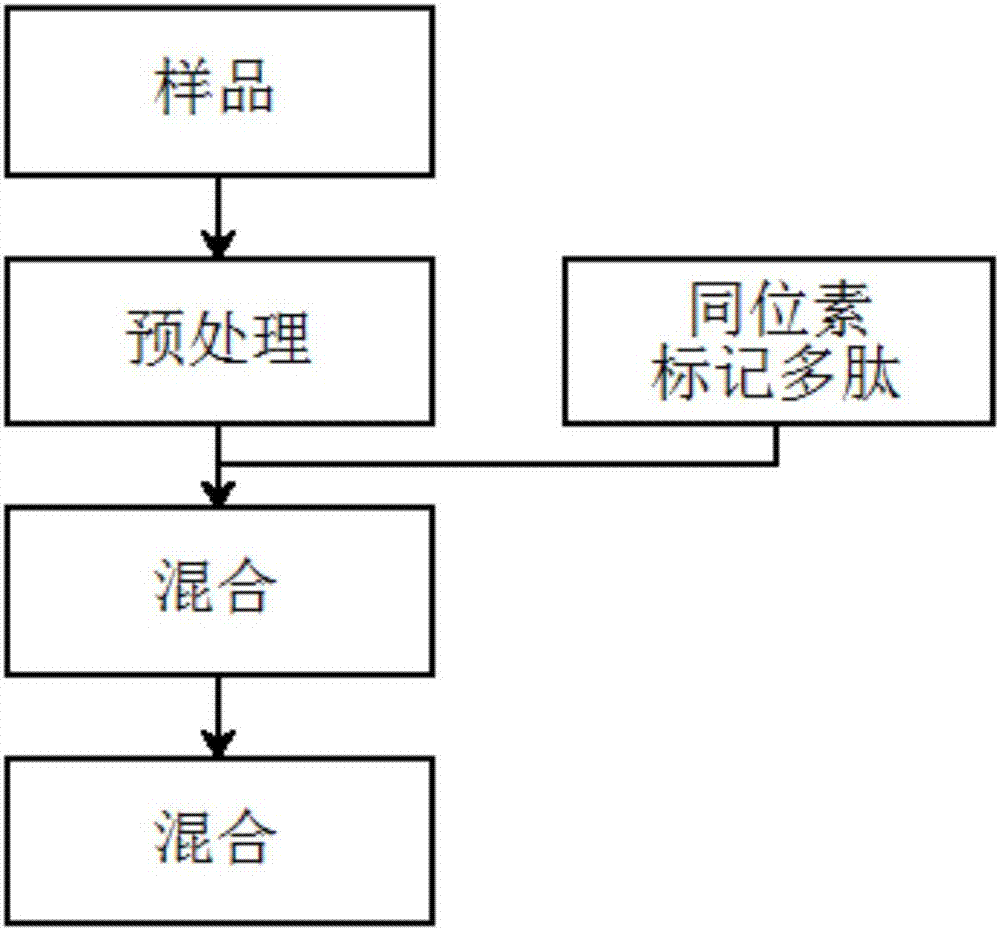
[0039] (1) Take 1 mL of red wine, add 100 μL of TEAB buffer (1M, pH=8.5), 10 μL of dithiothreitol (100 mmol / L), incubate in a water bath at 60°C for 30 minutes, add 10 μL of iodoacetamide ( 300mmol / L), at room temperature, keep it in the dark for 30 minutes, and obtain the labeled sample;
[0040] (2) Take the alpha s1-casein solution and process it in the same way as above, only replacing iodoacetamide with isotope-labeled 13 C 2 2 h 4 -Iodoacetamide, to obtain a labeled internal standard;
[0041] (3) Take 500 μL of the labeled sample and the labeled internal standard, mix them evenly, then add 10 μL of trypsin (100 μg / mL), and place them in a 37°C water bath for 4 hours to obtain the enzymatic hydrolysis solution;
[0042] (4) After the enzymolysis solution is filtered, the liq...
Embodiment 2
[0048] This embodiment is the detection of alpha s1-casein (milk allergen) in red wine, wherein the alpha s1-casein content is 20 μg / mL, and the detection process includes:
[0049] Take 1 mL of red wine, add 10 mL of acetonitrile to denature and precipitate the protein, centrifuge at 15,000 g for 30 minutes, discard the liquid, add 100 μL of TEAB buffer (1M, pH=8.5), 10 μL of RapiGest surfactant denaturant (1 mg / mL), Dithiothreitol 10 μL (100 mmol / L), water 990 μL, incubate in a water bath at 60° C. for 30 minutes; subsequent operations are the same as in Example 1 after “adding 10 μL iodoacetamide”.
Embodiment 3
[0065] Embodiment 3 (detection of beta lactoglobulin in protein drink)
[0066] In this embodiment, the steps for detecting beta lactoglobulin in protein drinks include:
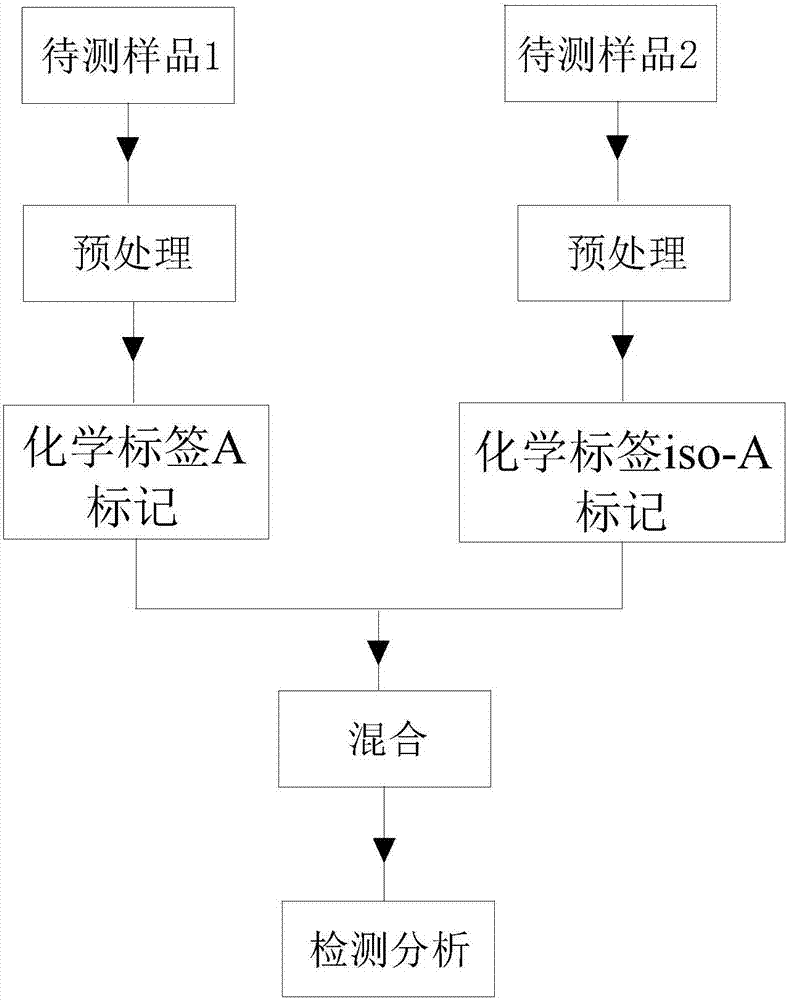
[0067] (1) Take 1mL beverage sample and beta lactoglobulin (20mg / L), add 100μL TEAB buffer (1M, pH=8.5) respectively, then add 40μL formaldehyde solution (4%) and 40μL sodium cyanoborohydride (0.6mol / L), after shaking and incubating at room temperature for 1 hour, the formaldehyde solution that reacts with the beverage sample is selected as non-isotope-labeled formaldehyde, and the formaldehyde solution that reacts with the beta lactoglobulin solution is 13 C 2 h 2 -formaldehyde;
[0068] (2) After shaking and incubating for 1 hour, add 160 μL ammonia water (1%) and 80 μL formic acid (5%) in sequence, then take 500 μL beverage sample solution and beta lactoglobulin solution respectively, mix and add 10 μL dithiothreitol ( 100mmol / L), after incubating in a water bath at 60°C for 30 minutes, add 10 μL (300...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com