PiRNA combination for detecting heart disease, and application thereof
A technology for heart disease and heart tissue, applied in the direction of DNA/RNA fragments, recombinant DNA technology, microbial determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
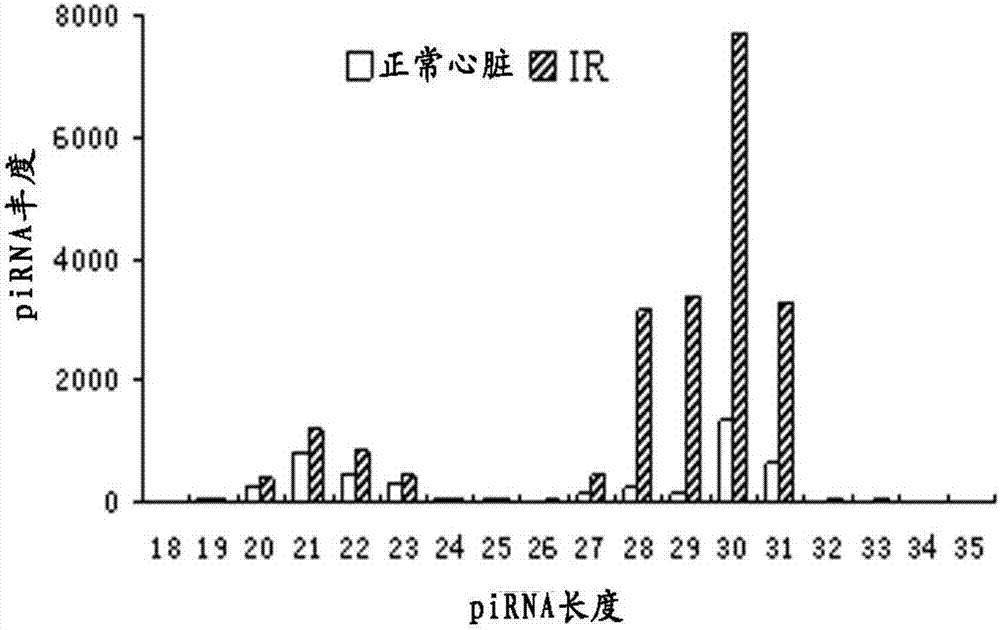
[0088] Example 1 Rapid screening of differentially expressed hearts under the ischemia-reperfusion model by high-throughput sequencing piRNA
[0089] Take C57 mature male mice more than 8 weeks old, and make myocardial infarction model according to the method of our laboratory, that is, the left anterior descending branch of the left ventricle of the mouse is ligated after thoracotomy to make the left ventricle ischemia for 45 minutes, then loosen the ligation, and reperfuse After 3 hours, observe and determine the infarction of the myocardium. The infarcted tissue and normal control heart tissue were quickly placed in liquid nitrogen and sent to Shenzhen Huada Gene Company for further RNA extraction, detection and sequencing. The sequencing method used is high-throughput Solexa deep sequencing, which can obtain a large amount of small RNA information in a short time. The small RNA (sRNA) obtained by Solexa deep sequencing covers almost all RNAs, including miRNA, siRNA, p...
Embodiment 2
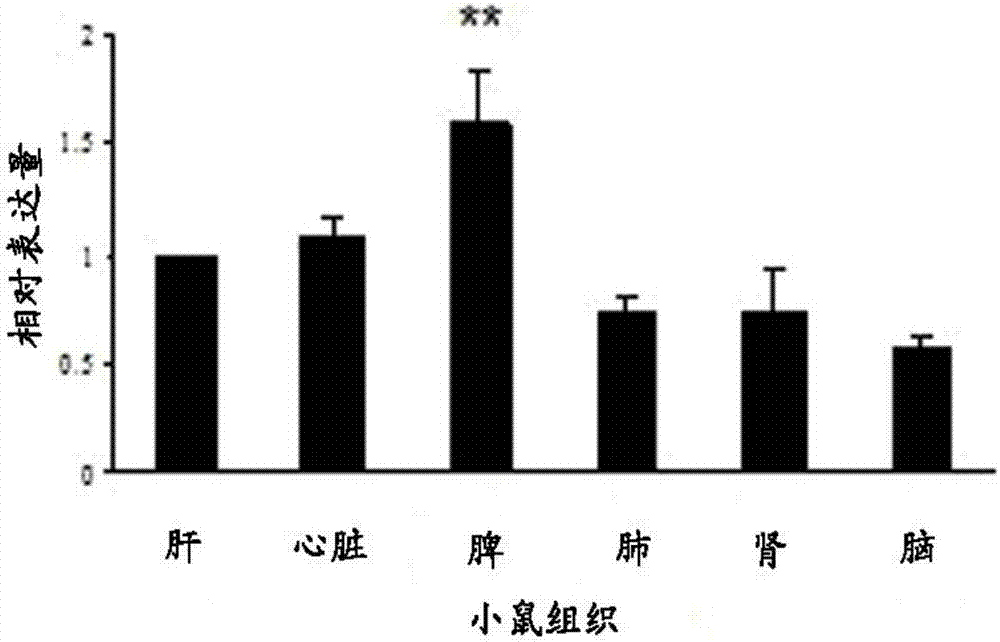
[0099] Example 2 Using fluorescent quantitative PCR method to detect the expression of piRNA in primary cardiomyocytes
[0100] The specific steps for the preparation of primary cardiomyocytes are as follows: the heart of a 2-day-old C57 mouse was peeled off, chopped and put into phosphate buffer (1L distilled water contained 8g of NaCl, 0.2g of KCl, anhydrous Na 2 HPO 4 1.44g, KH 2 PO 4 0.24g), then at 37°C, HEPES-buffered saline solution (20mM / L HEPES-NaOH, pH7.6, 130mM / L NaCl, 3mM / LKCl, 1mM / LNaH 2 PO 4 , 4mM / L glucose and 0.15% trypsin by weight / volume ratio), after centrifugation, the cells were resuspended in heat-inactivated horse serum containing 5% by volume, 100mM / L ascorbic acid, 1mg / mL transferrin, 10ng / mL Selenium, 100U / mL penicillin, 100U / mL streptomycin in DMEM / F12 cell culture medium. The isolated cardiomyocytes were cultured at 37°C for 1 hour, and then diluted to 1×10 6 The cells / mL were spread in a culture dish, and 0.1mM / L 5-bromodeoxyuridine was add...
Embodiment 3
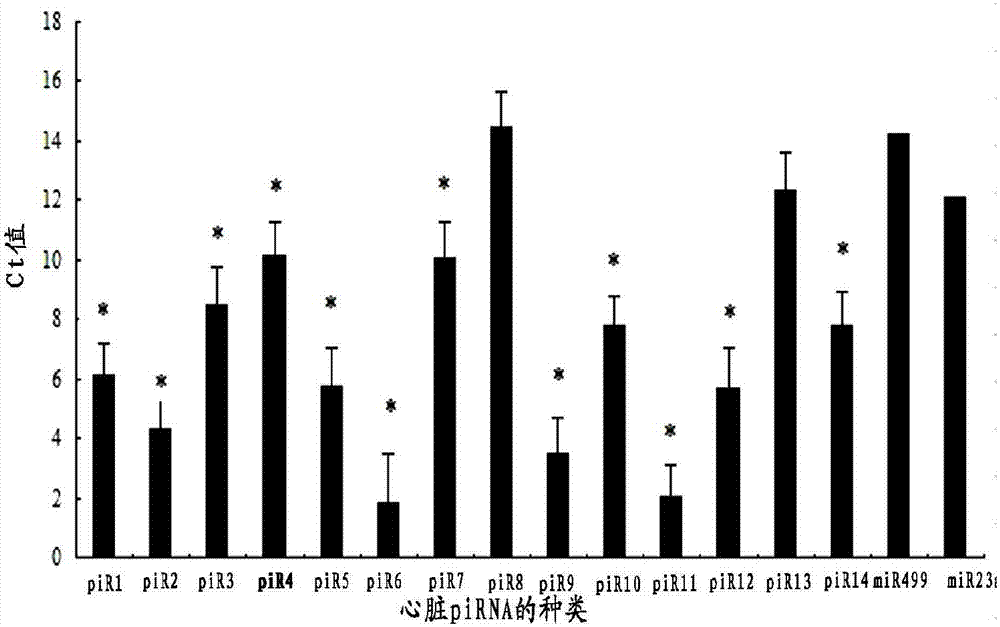
[0109] Example 3 Detection of piRNA Expression in Normal Heart and Myocardial Infarction Heart Using Fluorescent Quantitative PCR Condition
[0110] Total RNA was extracted from normal heart tissue and myocardial infarction heart tissue, and 1 μg of the extracted RNA was reverse-transcribed with the participation of RNA reverse transcriptase RTAce (Toyobo) using the corresponding reverse transcription primers (Table 5, RT-PH). Reverse transcription in a constant temperature water bath at 42°C for 1 hour, denaturation at 99°C for 5 minutes, see Example 2 for the specific reverse transcription process. The reverse transcription product was detected by fluorescent quantitative PCR using PH-F and loop-R in Table 5. See Example 2 for the specific process. Here we have optimized four piRNAs, SEQ ID NO7, SEQ ID NO12, SEQ ID NO 15, and SEQ ID NO16, namely piR-7, piR-12, piR-15, and piR-16, respectively, and the corresponding fluorescent PCR probes were respectively For SEQ ID NO1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com