Acidic high-temperature-resistant cellulase Cel5 and gene and application thereof
A technology of cellulase and high temperature resistance, applied in the field of genetic engineering, can solve the problems of poor thermal stability, inappropriate pH range, low expression level, etc., and achieve the effect of easy fermentation and production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Cloning of embodiment 1 cellulase coding gene cel5
[0052]Genomic DNA of Talaromyces leycettanus JCM12802 was extracted, and the total DNA of Talaromycesleycettanus was used as a template, and cel-F and cel-R were used as primers for PCR amplification (see Table 1). The PCR reaction parameters were: 95°C for 5min; 94°C for 30sec, 60°C for 30sec , 72°C for 30 sec, 30 cycles, a fragment of about 1400bp was obtained, which was recovered and sent to Ruibo Biotechnology Co., Ltd. for sequencing.
[0053] Table 1 Primers required for this experiment
[0054]
Embodiment 2
[0055] The acquisition of embodiment 2 cellulase cDNA
[0056] Extraction of total RNA from Talaromyces leycettanus JCM12802, using Oligo(dT) 20 and reverse transcriptase to obtain a strand of cDNA, and then use cel5-F and cel5-R (see Table 1) to amplify the single-stranded cDNA to obtain the cDNA sequence of cellulase. The amplified product is recovered and sent to Ruibo Sequencing Biotech Ltd.
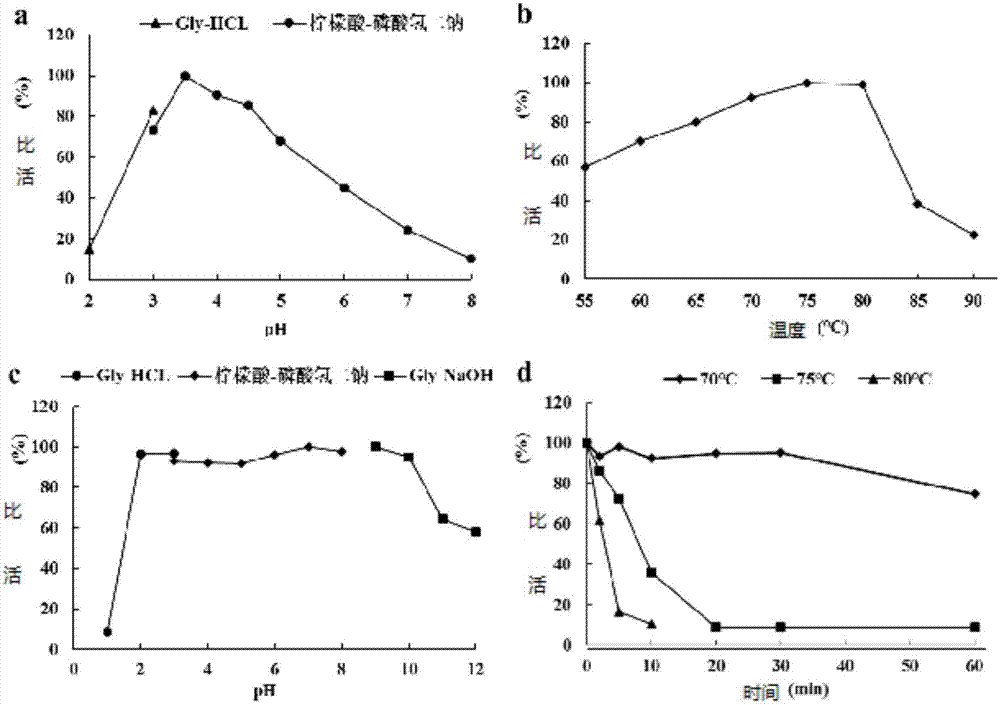
[0057] After comparing the genome sequence and cDNA sequence of cellulase, it was found that the gene contained 4 introns, the cDNA was 1230 bp long, encoded 409 amino acids and a stop codon, and the N-terminal 18 amino acids were its signal peptide sequence. The comparison proves that the cellulase-encoding gene isolated and cloned from Talaromyces leycettanus JCM12802 is a new gene.
Embodiment 3
[0058] The construction of embodiment 3 cellulase engineering strains
[0059] (1) Construction of expression vector and expression in yeast
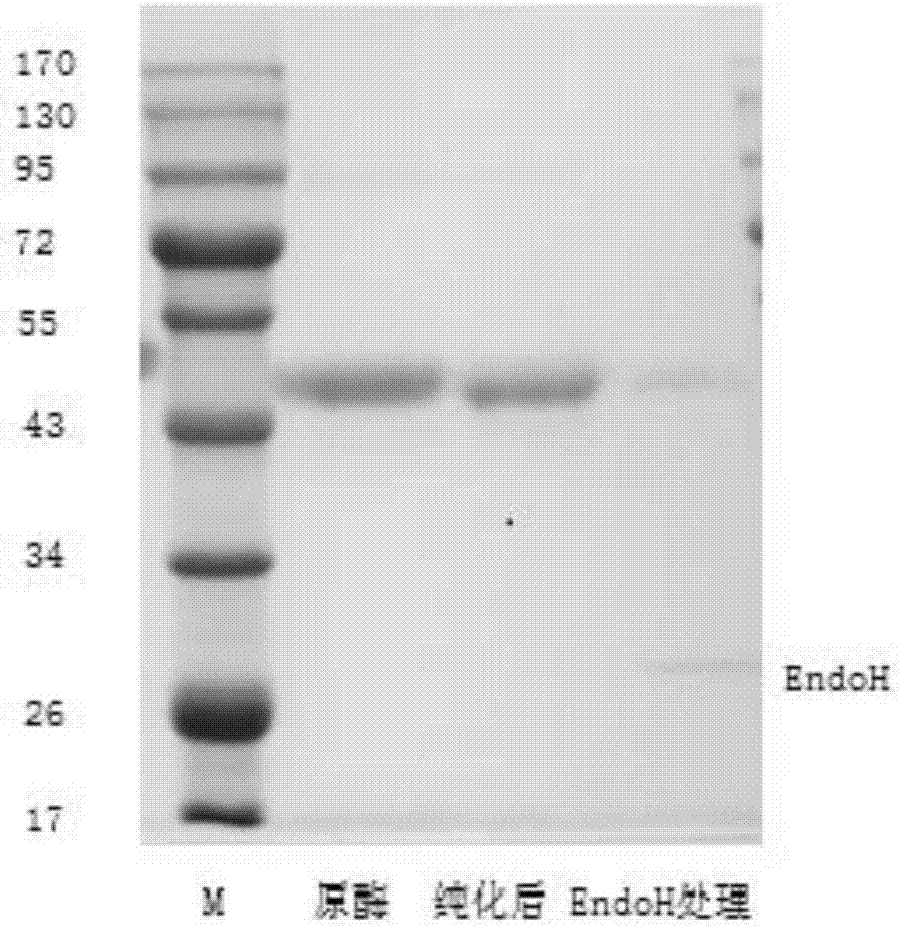
[0060] With the cDNA of cellulase cel5 correctly sequenced as a template, primers cel5-f and cel5-r (see Table 1) with SnaB I and Not I restriction enzyme sites were designed and synthesized, for the mature protein of Cel5 The coding region was amplified, and the PCR product was digested with SnaB I and Not I, and connected into the expression vector pPIC9 (Invitrogen, San Diego), and the sequence of the cellulase Cel5 mature protein was inserted into the downstream of the signal peptide sequence of the above expression vector, Form the correct reading frame with the signal peptide, construct the yeast expression vector pPIC9-cel5, and transform Escherichia coli competent cell Trans1. The positive transformants were subjected to DNA sequencing, and the transformants with the correct sequence were used for large-scale preparation of rec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com