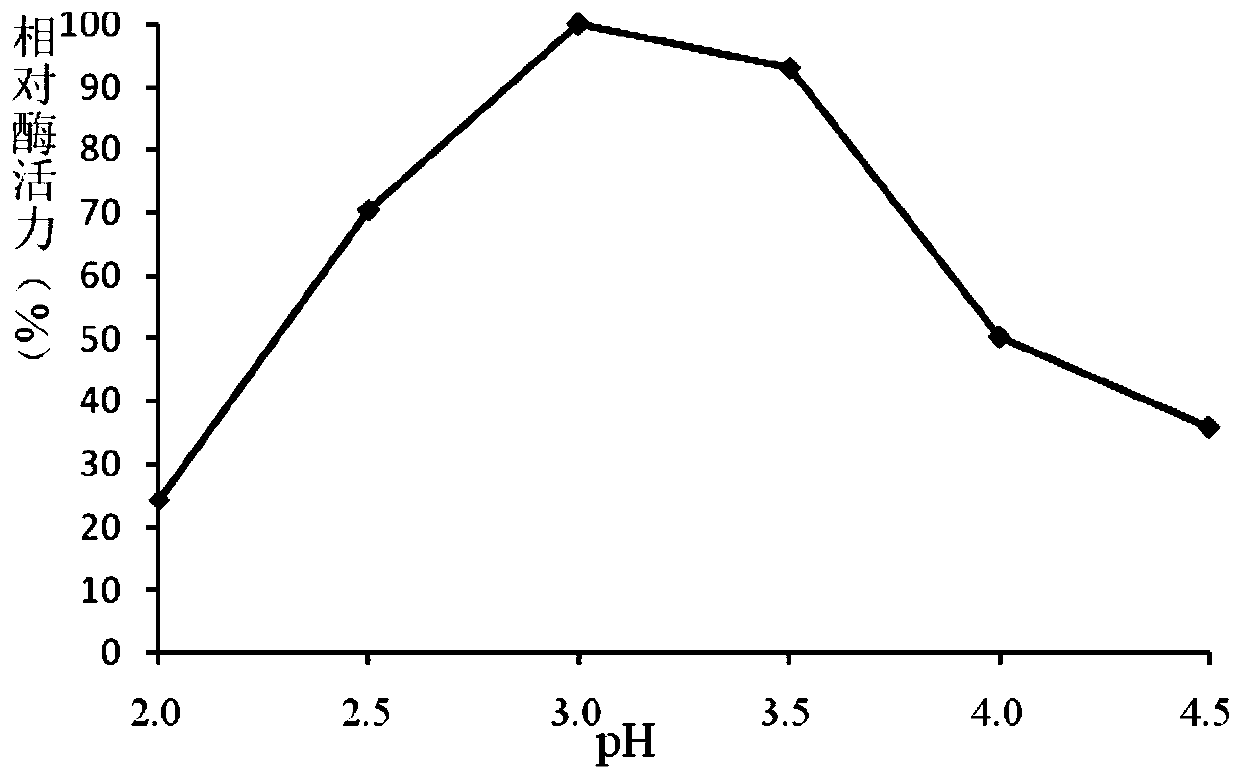
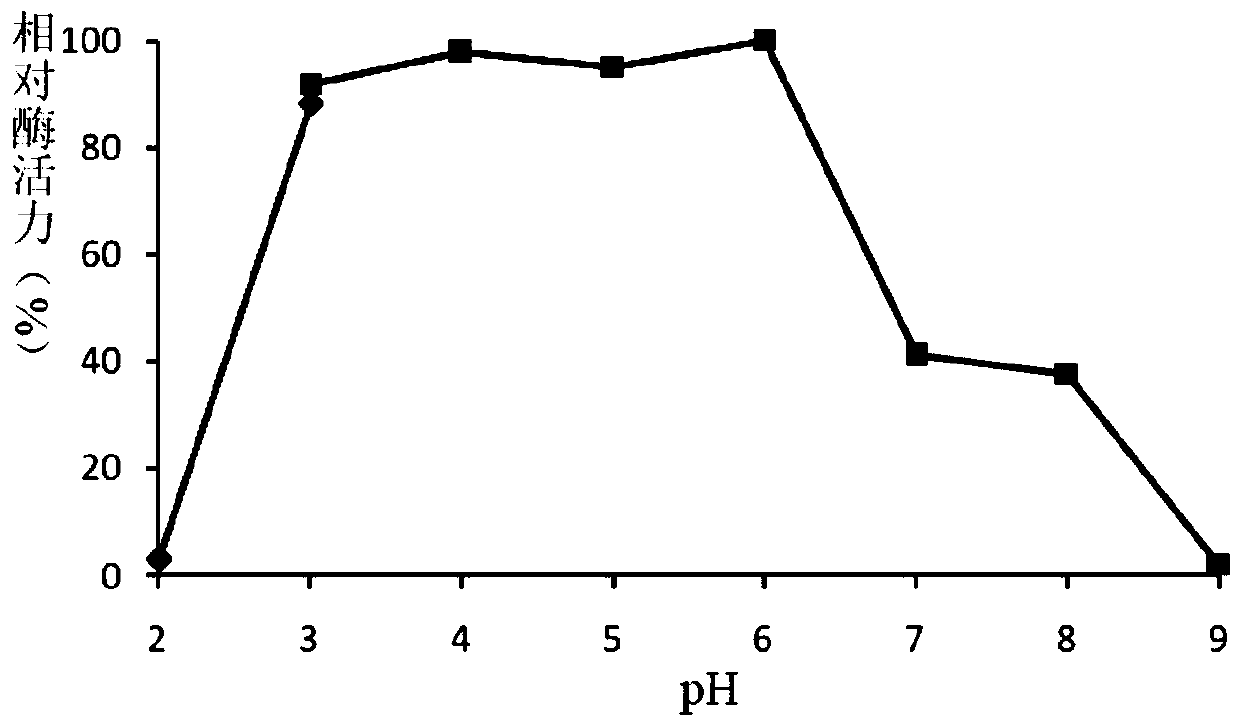
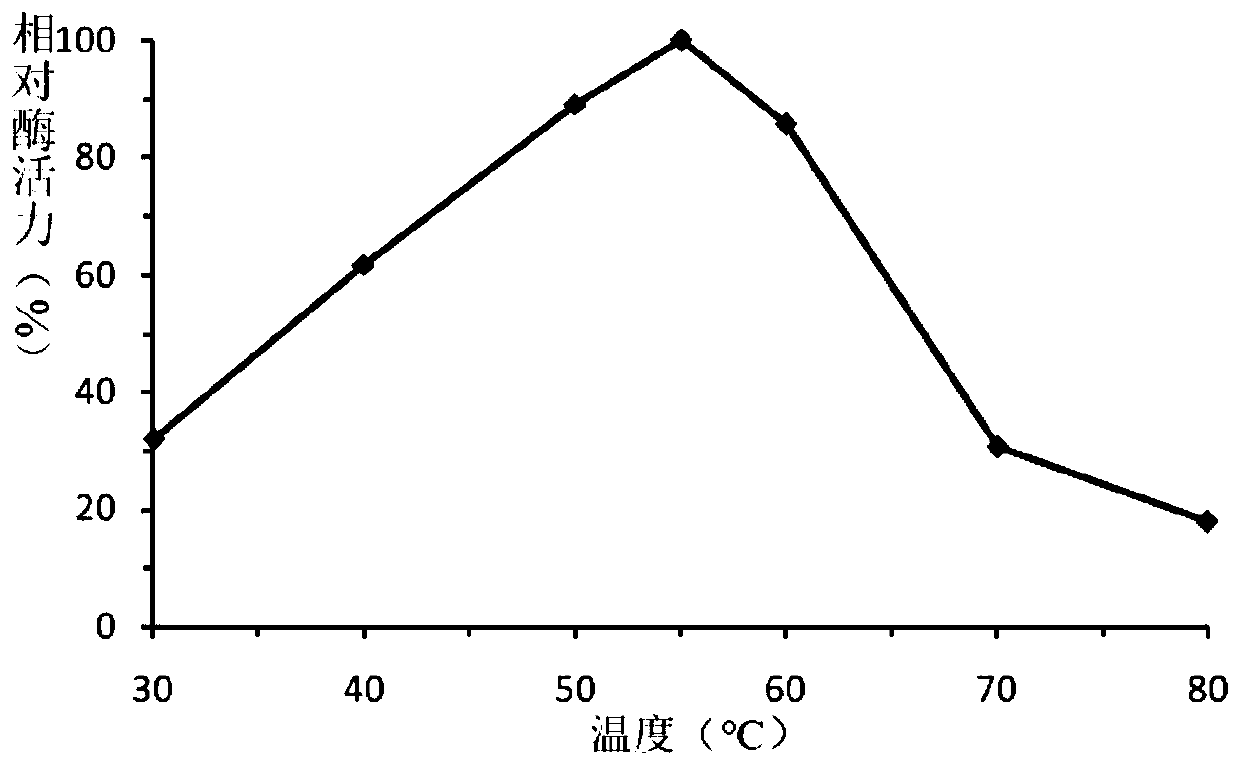
A fungus-derived acid protease g412 and its gene and application
An acid protease, ppic9-g412 technology, applied in the field of genetic engineering, can solve the problems of reduced catalytic efficiency, low enzymatic activity, industrial application limitations, etc., and achieves the effects of high reaction temperature and easy fermentation production.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Cloning of embodiment 1 protease coding gene g412
[0048] Genomic DNA of Talaromyces leycettanus JCM 12802 was extracted and stored at -20°C for later use.
[0049] Cloning primers g412F and g412R were designed, and the genomic DNA of Talaromyces leycettanus JCM 12802 was used as a template for PCR amplification. The PCR reaction parameters are: the PCR reaction parameters are: 95°C for 5min; 94°C for 30sec, 60°C for 30sec, 72°C for 2min, 35 cycles, and 72°C for 10min. A fragment of about 1800bp was obtained, which was recovered and sent to Ruibo Biotechnology Co., Ltd. for sequencing.
[0050] Table 1 Primers required for this experiment
[0051]
[0052] Extraction of total RNA from Talaromyces leycettanus JCM 12802, using Oligo(dT) 20and reverse transcriptase to obtain a strand of cDNA, then design the primers g412F and g412R (see Table 1) for amplifying the open reading frame, amplify the single-stranded cDNA, obtain the cDNA sequence of the protease, and sen...
Embodiment 2
[0054] The construction of embodiment 2 protease engineering strains
[0055] (1) Construction of expression vector and expression in yeast
[0056] Using the correctly sequenced cDNA of protease g412 as a template, primers F and R with SnaB I and Not I restriction sites were designed and synthesized (see Table 1) to amplify the coding region of the mature protein of g412. And use SnaB I and NotI to digest the PCR product, connect it into the expression vector pPIC9 (Invitrogen, San Diego), the sequence of the protease g412 mature protein is inserted into the downstream of the signal peptide sequence of the above expression vector, and form a correct reading frame with the signal peptide, The yeast expression vector pPIC9-g412 was constructed and transformed into Escherichia coli competent cell Trans1. The positive transformants were subjected to DNA sequencing, and the transformants with the correct sequence were used for large-scale preparation of recombinant plasmids. Lin...
Embodiment 3
[0060] The preparation of embodiment 3 recombinant proteases
[0061] (1) Massive expression of protease gene g412 at shake flask level in Pichia pastoris
[0062] The transformant with high enzyme activity was screened out, inoculated into a 1L Erlenmeyer flask with 300mL of BMGY liquid medium, cultured on a shaking table at 30°C at 220rpm for 48h; centrifuged at 5,000rpm for 5min, discarded the supernatant gently, and then added 100mL containing 0.5% methanol BMMY liquid medium, 30°C, 220rpm induction culture for 72h. During the induction culture period, add methanol solution once every 24 hours to compensate for the loss of methanol, and keep the methanol concentration at about 0.5%; (3) Centrifuge at 12,000×g for 10 minutes, collect the supernatant fermentation liquid, detect the enzyme activity and perform SDS-PAGE protein Electrophoretic analysis.
[0063] (2) Purification of recombinant protease
[0064] The recombinant protease supernatant expressed in the shake fla...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com