Method for detecting deficiency of 7q11.23
An analysis method and sequence technology, applied in the field of molecular biology, to achieve the effect of reducing operation steps, high accuracy and shortening detection cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Embodiment 1 detection method
[0028] 1. Materials and Instruments
[0029] Rotor-Gene Q real-time fluorescent quantitative PCR analyzer (qiagen), Klentaq enzyme (Ab Peptides), LCgreen fluorescent dye (BioFire Defense), primers (Shanghai Sangong Biotechnology), blood DNA extraction samples from patients with Williams–Beuren syndrome (7q11. 23 long fragment deletion), healthy human blood DNA extraction samples (no deletion of 7q11.23).
[0030] 2. Method
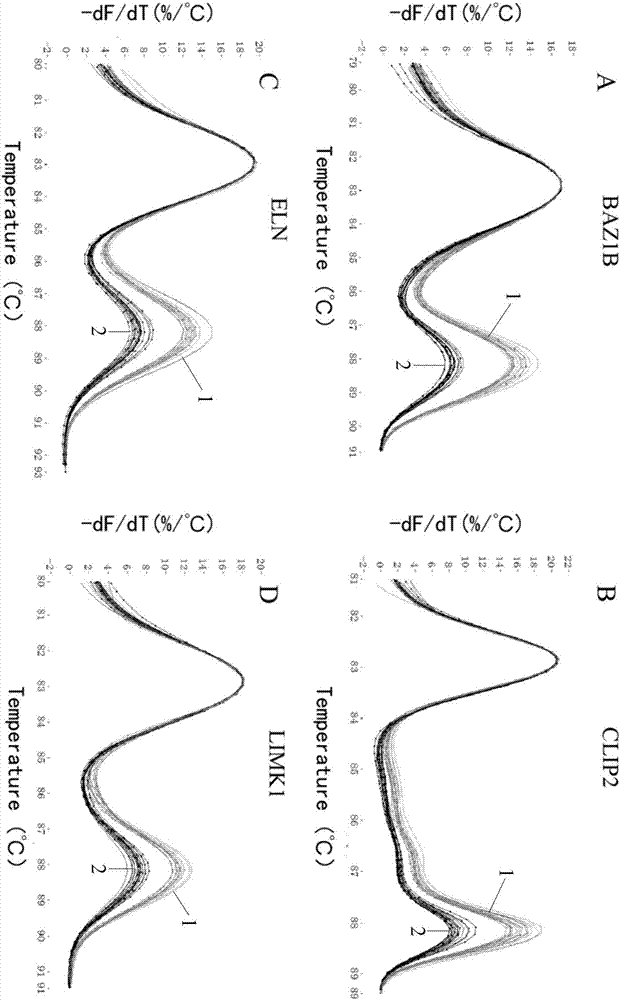
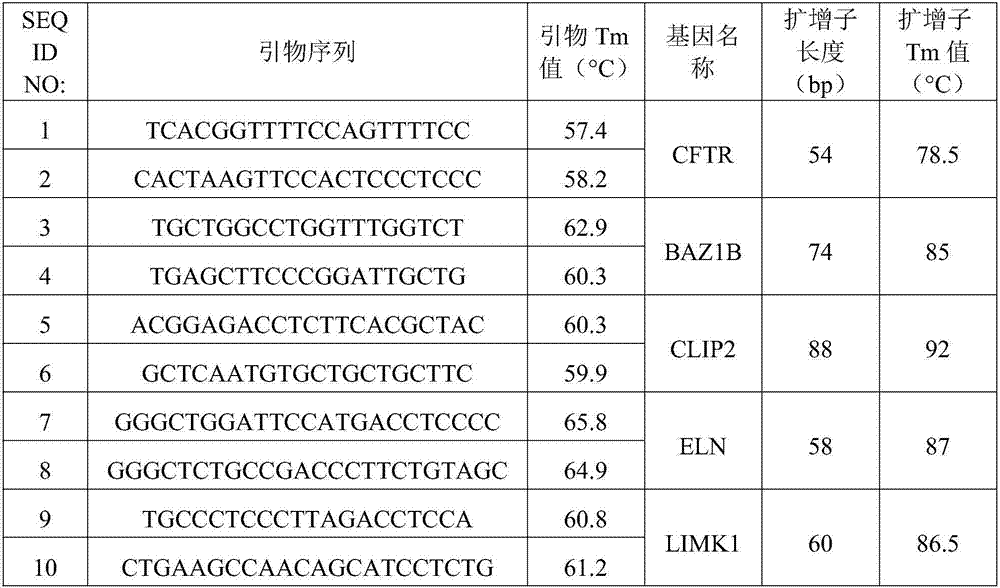
[0031] 1. Primer design: Four genes were selected in the deletion-prone region of 7q11.23, which were located at the proximal end of the region (BAZ1B), near the centromere end (CLIP2) and in the middle (ELN, LIMK1). Primers were designed for these four genes as the target genes, so that they can specifically amplify the amplicons of 58-88bp. At the same time, primers were designed using the stable 2-copy gene CFTR outside the 7q11.23 region as a control gene, and the length of the amplicon was 54bp. The Tm values...
Embodiment 2
[0039] Verification of embodiment 2 detection method
[0040] The detection method of Example 1 and the Affymetrix gene chip were used to detect and analyze the blood DNA extraction samples of unknown 7q11.23 deletion, and clinical observation was carried out. The results were compared, and the accuracy rate was 100%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com