A method for identifying adelphocoris ngritylus populations by utilizing molecular markers
A technology of Lygus sativa and black lip, applied in the direction of DNA / RNA fragments, recombinant DNA technology, etc., can solve problems such as difficult identification, and achieve strong operability, short time-consuming, and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Design and synthesis of PCR primer combinations for identification of Lygus black-lipped populations
[0039] Based on the mitochondrial cytochrome oxidase I (COI) gene and the mitochondrial NADH dehydrogenase subunit 4 (ND4) gene of Lygus alfalfa, the Primer Primer5 software was used to design specific primers for PCR amplification of the two genes. The primer sequences are as follows (SEQ ID NOs: 1-4):
[0040] ①Ngri-COI-F:
[0041] 5'-ATGAATAAATGATTATTTTCCACAAATCA-3' (SEQ ID NO. 1)
[0042] Ngri-COI-R:
[0043] 5'-CTGAATAATTAATATTTGTGCCATGAATG-3' (SEQ ID NO. 2)
[0044] ②Ngri-ND4-F:
[0045] 5'-AAAATACATTAAAGTAAACAAATCAATTT-3' (SEQ ID NO. 3)
[0046] Ngri-ND4-R:
[0047] 5'-ATATATTATTTATTTTTTGAGTGTAGTTTAATCCCT-3' (SEQ ID NO. 4)
[0048] The above primers were synthesized by Shanghai Bioengineering Technology Service Co., Ltd.
Embodiment 2
[0049] Embodiment 2 Utilize molecular markers to quickly identify the method for Northeast population and North China population of Lygus californica
[0050] 1. Collect the individuals of Lygus black-lipped alfalfa in Northeast China and North China, and extract genomic DNA
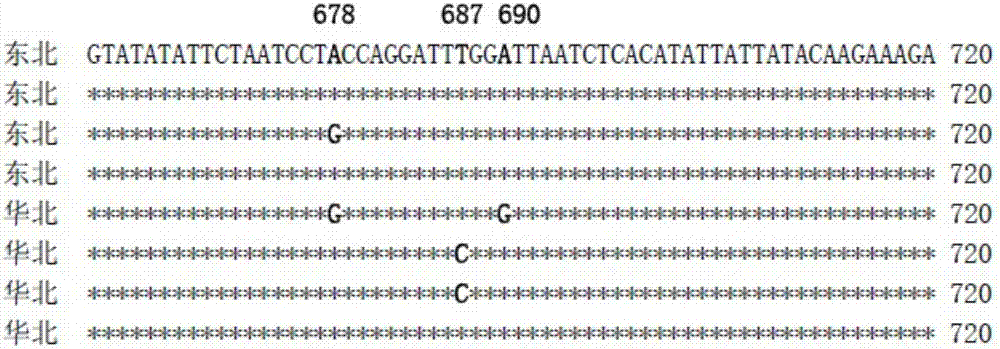
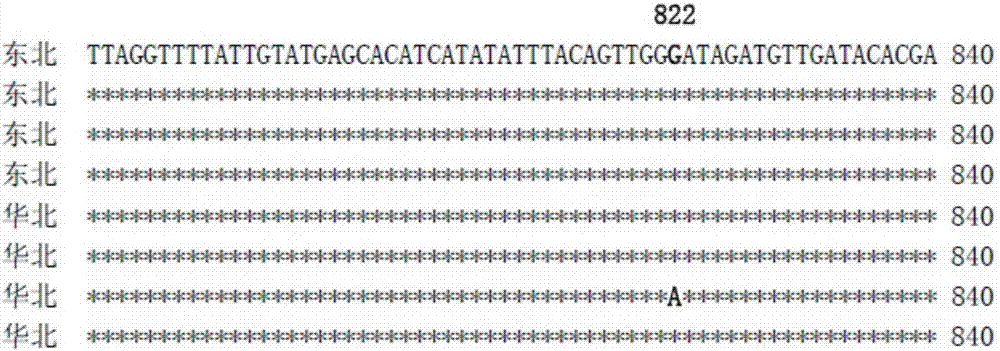
[0051] Representative populations were selected from Northeast China and North China, namely, Tieling City in Liaoning Province and Suihua City in Heilongjiang Province in Northeast China, Zhengzhou City in Henan Province, Hengshui City in Hebei Province and Dezhou City in Shandong Province in North China. . The field collection of Lygus black-lipped alfalfa adopts the net capture method. The adults are collected through a fluke tube, and then the adults are placed in analytically pure ethanol, and then brought back to the laboratory. The asana dissection microscope is used to refer to the relevant literature such as "Chinese Zoology" and so on. Further identification and confirmation of the external mo...
Embodiment 3
[0064] Example 3 Analysis of Specificity and Sensitivity of Detecting Lygus alfalfa by PCR Primer Ngri-COI-F / Ngri-COI-R
[0065] 1. Source of samples: In this example, Lygus pratensis (L.) was used as a control, and Lygus alfalfa came from Tieling, Liaoning in Northeast, Suihua in Heilongjiang and Zhengzhou in Henan, Hengshui in Hebei, and Dezhou in Shandong in North China; samples were used. It is preserved in the Plant Protection Laboratory, Cotton Research Institute, Chinese Academy of Agricultural Sciences.
[0066] 2. DNA extraction: as described in Example 2.
[0067] 3. PCR amplification: The PCR reaction system and PCR amplification procedure are the same as those described in Example 2.
[0068] 4. Result analysis: Take 5 μL of PCR amplification product, use 2% agarose gel for electrophoresis, the voltage is 120V, and test the result under ultraviolet light after 25 to 30 minutes. If there is a characteristic band of about 1100bp in size, it proves that The tested s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com