Ring mediated cascade amplification strategy used for detecting DNA transmethylase activity in high sensitivity manner
A methyltransferase and activity technology, applied in the field of enzyme activity detection, can solve the problems of non-specific background amplification, probe leakage non-specific amplification, affecting detection sensitivity and accuracy, etc. selective effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] 1. Analysis of Dam methyltransferase activity
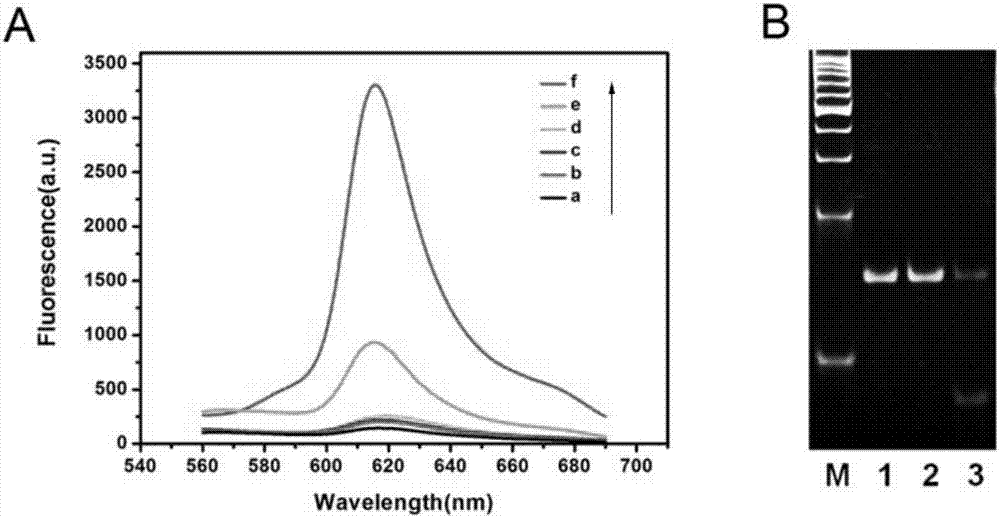
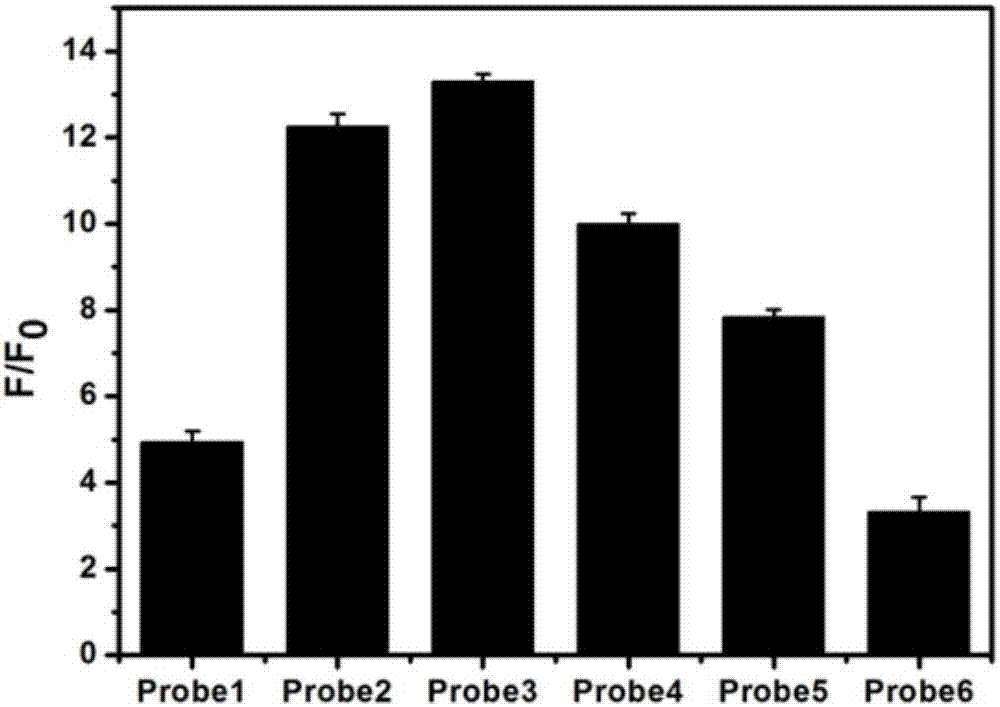
[0068] To 10 μL methyltransferase buffer (50mM NaCl, 10mM Tris-HCl, 10mM MgCl 2 , 1mMdithiothreitol, pH 7.5) were added with 160μM SAM (SAM is a cofactor), 20nM long stem-loop probe, 2U DpnI and different amounts of Dam methyltransferase, and incubated at 37°C for 2h. Subsequently, 40nM hairpin probe, 1UKlenow Fragment (3′–5′exo-), 2U Nt.BbvCI and 0.6mM dNTPs and 1×CutSmart (50mM KAc, 20mM Tris-HAc, 10mM Mg( Ac) 2 , 100μg / mL BSA, pH 7.9), incubate at 37°C for 30 min. The reaction system was heated at 85 °C for 20 min to inactivate the enzyme, and then slowly cooled to 30 °C. After that, 120U T4 DNA ligase, 800nM padlock probe and 1×T4 ligase buffer (50mM Tris-HCl, 10mM MgCl 2 , 10mMdithiothreitol, 1mM ATP, pH 7.5), incubated at 37°C for 1h. Then 1mM dNTPs, 3U Phi29 DNA polymerase, 4U Nt.BbvCI and 1×CutSmart were added, and incubated at 37°C for 3h. The reaction system was heated at 75 °C for 20 min to inactivate the ...
Embodiment 2
[0074] 1. Principle analysis
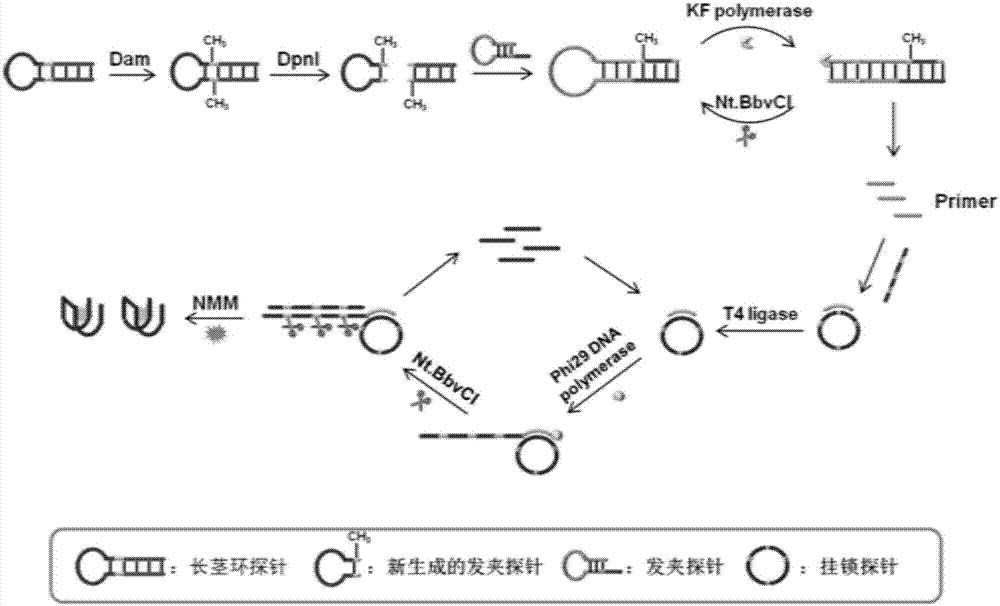
[0075] The proposed rationale for the assay of methyltransferase activity is as follows: figure 1shown. When Dam methyltransferase exists, it specifically recognizes and catalyzes the methylation of long stem-loop probes to form methylated long stem-loops. Subsequently, the DpnI restriction enzyme specifically cuts the methylated long stem-loop into two parts. One part is double-stranded DNA, and the other part is new hairpin probe. Under the experimental conditions, the structure of the new hairpin probe is unstable, and a conformational transition occurs to form a single strand. Under the action of Klenow Fragment polymerase and Nt.BbvCI, the single-stranded DNA initiates polymerization and cleavage reactions, releasing multiple primers. Under the action of T4 DNA ligase, the released primer hybridizes with the padlock probe to obtain a circular probe. Subsequently, this primer triggers rolling circle amplification under the action of Phi2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com