Multiple detection method for field screening of toxicogenic Pseudomonas aeruginosa
A Pseudomonas aeruginosa, multiple detection technology, applied in the field of molecular biology, can solve the problems that the analysis and identification of amplification products need to be improved, and the identification of molecular weight target products cannot be distinguished, and achieves shortened detection time, high sensitivity, and improved detection efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] The concrete detection method of the present embodiment is:
[0038] 1. Extraction of sample DNA
[0039] 1.1 The standard strains of Pseudomonas aeruginosa were inoculated in the nutrient broth and cultured at 36°С±1°С for 18 hours.
[0040] 1.2 Use the bacterial genome DNA extraction kit to extract the DNA of the above culture products respectively, and use it as a template for detection.
[0041] 1.3 Mix the two sets of DNA extracts in an equal volume ratio as a template for the detection of mixed target bacteria.
[0042] 2. Multiple LAMP reactions
[0043] 2.1 Artificially synthesized labeled primer sets for the internal standard gene and two toxin genes of Pseudomonas aeruginosa respectively.
[0044] 2.2 Multiple LAMP reaction system
[0045] The composition of the reaction system includes: (1) primer mixture: outer primer F 3 and B 3 0.3 μmol / L each, 2.3 μmol / L each for internal primers FIP and BIP; (2) Reaction mixture: 3mol / L betaine, 0.3mmol / L MgSO 4 , 1...
Embodiment 2
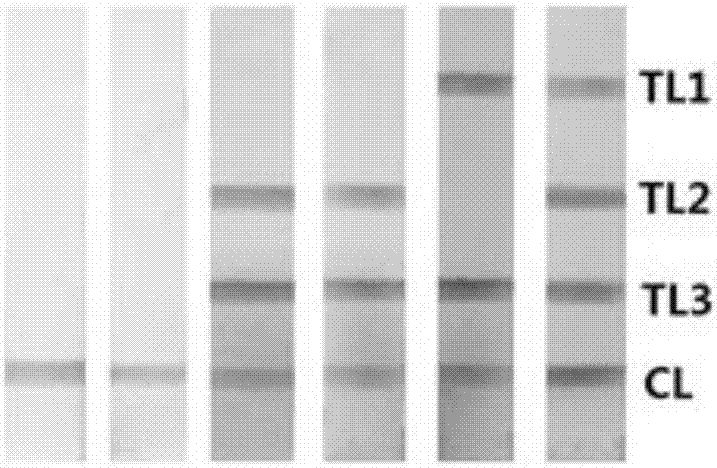
[0057] Take 63 samples of river water, take 2ml of sample solution to extract the genome and perform multiple LAMP amplification, then use colloidal gold test strips to detect multiple LAMP products, and read the test results within 5 minutes. In the case of color development of the quality control line: the detection lines T3, T2 and T1 are not colored, indicating that there is no Pseudomonas aeruginosa in the sample; the detection line T3 is colored, and T2 and T1 are not colored, indicating that there is no color in the sample There are Pseudomonas aeruginosa that does not secrete toxin ExoU and ExoS genes; detection lines T3 and T2 develop color, and T1 does not develop color, indicating that there are Pseudomonas aeruginosa that secrete toxin ExoS but not ExoU genes in the sample; detection line T3 and T1 color development, T2 no color development, indicating that there is Pseudomonas aeruginosa that secretes the toxin ExoU but not the ExoS gene in the sample; the detectio...
Embodiment 3
[0059] Take 78 samples of drinking water, take 2ml of sample solution to extract the genome and perform multiple LAMP amplification, then use colloidal gold test strips to detect multiple LAMP products, and read the test results within 5 minutes. In the case of color development of the quality control line: the detection lines T3, T2 and T1 are not colored, indicating that there is no Pseudomonas aeruginosa in the sample; the detection line T3 is colored, and T2 and T1 are not colored, indicating that there is no color in the sample There are Pseudomonas aeruginosa that does not secrete toxin ExoU and ExoS genes; detection lines T3 and T2 develop color, and T1 does not develop color, indicating that there are Pseudomonas aeruginosa that secrete toxin ExoS but not ExoU genes in the sample; detection line T3 and T1 color development, T2 no color development, indicating that there is Pseudomonas aeruginosa that secretes the toxin ExoU but not the ExoS gene in the sample; the detec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com